How diverse is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae)
- PMID: 19098217
- PMCID: PMC2643572
- DOI: 10.1128/AEM.01109-08
How diverse is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae)
Abstract
At least 20% of all arthropods and some nematode species are infected with intracellular bacteria of the genus Wolbachia. This highly diverse genus has been subdivided into eight "supergroups" (A to H) on the basis of nucleotide sequence data. Here, we report the discovery of a new Wolbachia supergroup recovered from the spider mite species Bryobia species V (Acari: Tetranychidae), based on the sequences of three protein-coding genes (ftsZ, gltA, and groEL) and the 16S rRNA gene. Other tetranychid mites possess supergroup B Wolbachia strains. The discovery of another Wolbachia supergroup expands the known diversity of Wolbachia and emphasizes the high variability of the genus. Our data also clarify the existing supergroup structure and highlight the use of multiple gene sequences for robust phylogenetic analysis. In addition to previous reports of recombination between the arthropod-infecting supergroups A and B, we provide evidence for recombination between the nematode-infecting supergroups C and D. Robust delineation of supergroups is essential for understanding the origin and spread of this common reproductive parasite and for unraveling mechanisms of host adaptation and manipulation across a wide range of hosts.
Figures

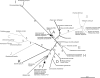

Comment in
-
Recombination in wolbachia endosymbionts of filarial nematodes?Appl Environ Microbiol. 2011 Mar;77(5):1921-2. doi: 10.1128/AEM.02380-10. Appl Environ Microbiol. 2011. PMID: 21350051 Free PMC article. No abstract available.
References
-
- Akaike, H. 1974. A new look at the statistical model identification. IEEE Trans. Autom. Contr. 19:716-723.
-
- Andersson, S. G. E., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. M. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. - PubMed
-
- Baldo, L., S. Bordenstein, J. J. Wernegreen, and J. H. Werren. 2006. Widespread recombination throughout Wolbachia genomes. Mol. Biol. Evol. 23:437-449. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

