Genetic diversity of native bradyrhizobia isolated from soybeans (Glycine max L.) in different agricultural-ecological-climatic regions of India
- PMID: 18676699
- PMCID: PMC2565974
- DOI: 10.1128/AEM.01320-08
Genetic diversity of native bradyrhizobia isolated from soybeans (Glycine max L.) in different agricultural-ecological-climatic regions of India
Abstract
Fifty isolates from root nodules of soybean plants sampled in five agricultural-ecological-climatic regions of India were analyzed by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene, the intergenic spacer region between the 16S and 23S rRNA genes (IGS), and the nifH and nodC genes. Eight haplotypes assigned to the Bradyrhizobium genus were identified, and the genetic diversity was conserved across regions. Sequence analyses of the IGS and the dnaK, glnII, recA, and nifH genes revealed three groups. One of them (26% of isolates) was assigned to Bradyrhizobium liaoningense. A second group (36% of isolates) was identified as B. yuanmingense but likely forms a new biovar able to nodulate soybean plants. The third lineage (38% of isolates) was different from all described Bradyrhizobium species but showed the same symbiotic genotype as B. liaoningense and B. japonicum bv. glycinearum.
Figures

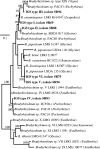

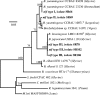
References
-
- Barcellos, F. G., P. Menna, J. S. da Silva Batista, and M. Hungria. 2007. Evidence of horizontal transfer of symbiotic genes from a Bradyrhizobium japonicum inoculant strain to indigenous diazotrophs Sinorhizobium (Ensifer) fredii and Bradyrhizobium elkanii in a Brazilian Savannah soil. Appl. Environ. Microbiol. 73:2635-2643. - PMC - PubMed
-
- Chauhan, G. S., and O. P. Joshi. 2005. Soybean (Glycine max)—the 21st century crop. Indian J. Agric. Sci. 75:461-469.
-
- Chen, W., E. Wang, S. Wang, Y. Li, and X. Chen. 1995. Characteristics of Rhizobium tianshanense sp. nov., a moderately and slowly growing root nodule bacterium isolated from an arid saline environment in Xinjiang, People's Republic of China. Int. J. Syst. Bacteriol. 45:153-159. - PubMed
-
- Chen, W. X., G. H. Yan, and J. L. Li. 1988. Numerical taxonomic study of fast-growing soybean rhizobia and a proposal that Rhizobium fredii be assigned to Sinorhizobium gen. nov. Int. J. Syst. Bacteriol. 38:392-397.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

