Genome variation and population structure among 1142 mosquitoes of the African malaria vector species Anopheles gambiae and Anopheles coluzzii
- PMID: 32989001
- PMCID: PMC7605271
- DOI: 10.1101/gr.262790.120
Genome variation and population structure among 1142 mosquitoes of the African malaria vector species Anopheles gambiae and Anopheles coluzzii
Abstract
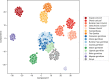
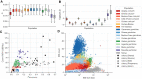
Mosquito control remains a central pillar of efforts to reduce malaria burden in sub-Saharan Africa. However, insecticide resistance is entrenched in malaria vector populations, and countries with a high malaria burden face a daunting challenge to sustain malaria control with a limited set of surveillance and intervention tools. Here we report on the second phase of a project to build an open resource of high-quality data on genome variation among natural populations of the major African malaria vector species Anopheles gambiae and Anopheles coluzzii We analyzed whole genomes of 1142 individual mosquitoes sampled from the wild in 13 African countries, as well as a further 234 individuals comprising parents and progeny of 11 laboratory crosses. The data resource includes high-confidence single-nucleotide polymorphism (SNP) calls at 57 million variable sites, genome-wide copy number variation (CNV) calls, and haplotypes phased at biallelic SNPs. We use these data to analyze genetic population structure and characterize genetic diversity within and between populations. We illustrate the utility of these data by investigating species differences in isolation by distance, genetic variation within proposed gene drive target sequences, and patterns of resistance to pyrethroid insecticides. This data resource provides a foundation for developing new operational systems for molecular surveillance and for accelerating research and development of new vector control tools. It also provides a unique resource for the study of population genomics and evolutionary biology in eukaryotic species with high levels of genetic diversity under strong anthropogenic evolutionary pressures.
© 2020 The Anopheles gambiae 1000 Genomes Consortium; Published by Cold Spring Harbor Laboratory Press.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
