Analysis of Systemic Epigenetic Alterations in Inflammatory Bowel Disease: Defining Geographical, Genetic and Immune-Inflammatory influences on the Circulating Methylome
- PMID: 36029471
- PMCID: PMC10024547
- DOI: 10.1093/ecco-jcc/jjac127
Analysis of Systemic Epigenetic Alterations in Inflammatory Bowel Disease: Defining Geographical, Genetic and Immune-Inflammatory influences on the Circulating Methylome
Abstract
Background: Epigenetic alterations may provide valuable insights into gene-environment interactions in the pathogenesis of inflammatory bowel disease [IBD].
Methods: Genome-wide methylation was measured from peripheral blood using the Illumina 450k platform in a case-control study in an inception cohort (295 controls, 154 Crohn's disease [CD], 161 ulcerative colitis [UC], 28 IBD unclassified [IBD-U)] with covariates of age, sex and cell counts, deconvoluted by the Houseman method. Genotyping was performed using Illumina HumanOmniExpressExome-8 BeadChips and gene expression using the Ion AmpliSeq Human Gene Expression Core Panel. Treatment escalation was characterized by the need for biological agents or surgery after initial disease remission.
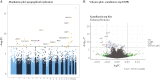
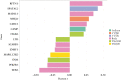
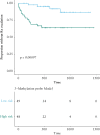
Results: A total of 137 differentially methylated positions [DMPs] were identified in IBD, including VMP1/MIR21 [p = 9.11 × 10-15] and RPS6KA2 [6.43 × 10-13], with consistency seen across Scandinavia and the UK. Dysregulated loci demonstrate strong genetic influence, notably VMP1 [p = 1.53 × 10-15]. Age acceleration is seen in IBD [coefficient 0.94, p < 2.2 × 10-16]. Several immuno-active genes demonstrated highly significant correlations between methylation and gene expression in IBD, in particular OSM: IBD r = -0.32, p = 3.64 × 10-7 vs non-IBD r = -0.14, p = 0.77]. Multi-omic integration of the methylome, genome and transcriptome also implicated specific pathways that associate with immune activation, response and regulation at disease inception. At follow-up, a signature of three DMPs [TAP1, TESPA1, RPTOR] were associated with treatment escalation to biological agents or surgery (hazard ratio of 5.19 [CI: 2.14-12.56], logrank p = 9.70 × 10-4).
Conclusion: These data demonstrate consistent epigenetic alterations at diagnosis in European patients with IBD, providing insights into the pathogenetic importance and translational potential of epigenetic mapping in complex disease.
Keywords: DNA methylation; Mendelian randomization; epigenetic clock; gene expression; genetics; inflammatory bowel diseases [IBD]; methylation; prognosis; quantitative trait loci.
© The Author(s) 2022. Published by Oxford University Press on behalf of European Crohn’s and Colitis Organisation.
Conflict of interest statement
R. Kalla Financial support for research: EC IBD-Character, Lecture fee[s]: Ferring, N. Kennedy Financial support for research: Wellcome Trust, Conflict with: Abbvie, MSD, Warner Chilcott, Ferring speaker fees. Shire Travel bursary, A. Adams: none declared, J. Satsangi Financial support for research: EC grant IBD-BIOM,IBD-Character, Wellcome, CSO, MRC, Conflict with: Consultant for: Takeda, Conflict with: MSD speaker fees. Shire travelling expenses
Figures

Comment in
-
Validation of IBD-associated Whole-blood DNA Methylation Changes-Where Do We Go From Here?J Crohns Colitis. 2023 Mar 18;17(2):151-152. doi: 10.1093/ecco-jcc/jjac163. J Crohns Colitis. 2023. PMID: 36367402 No abstract available.
References
-
- Cedar H, Bergman Y.. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 2009;10:295–304. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

