Subsurface cycling of nitrogen and anaerobic aromatic hydrocarbon biodegradation revealed by nucleic Acid and metabolic biomarkers
- PMID: 20348302
- PMCID: PMC2869145
- DOI: 10.1128/AEM.00172-10
Subsurface cycling of nitrogen and anaerobic aromatic hydrocarbon biodegradation revealed by nucleic Acid and metabolic biomarkers
Abstract
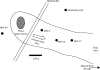
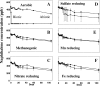
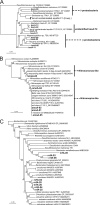
Microbial processes are crucial for ecosystem maintenance, yet documentation of these processes in complex open field sites is challenging. Here we used a multidisciplinary strategy (site geochemistry, laboratory biodegradation assays, and field extraction of molecular biomarkers) to deduce an ongoing linkage between aromatic hydrocarbon biodegradation and nitrogen cycling in a contaminated subsurface site. Three site wells were monitored over a 10-month period, which revealed fluctuating concentrations of nitrate, ammonia, sulfate, sulfide, methane, and other constituents. Biodegradation assays performed under multiple redox conditions indicated that naphthalene metabolism was favored under aerobic conditions. To explore in situ field processes, we measured metabolites of anaerobic naphthalene metabolism and expressed mRNA transcripts selected to document aerobic and anaerobic microbial transformations of ammonia, nitrate, and methylated aromatic contaminants. Gas chromatography-mass spectrometry detection of two carboxylated naphthalene metabolites and transcribed benzylsuccinate synthase, cytochrome c nitrite reductase, and ammonia monooxygenase genes indicated that anaerobic metabolism of aromatic compounds and both dissimilatory nitrate reduction to ammonia (DNRA) and nitrification occurred in situ. These data link formation (via DNRA) and destruction (via nitrification) of ammonia to in situ cycling of nitrogen in this subsurface habitat, where metabolism of aromatic pollutants has led to accumulation of reduced metabolic end products (e.g., ammonia and methane).
Figures
References
-
- Anneser, B., F. Einsiedl, R. U. Meckenstock, L. Richters, F. Wisotzky, and C. Griebler. 2008. High-resolution monitoring of biogeochemical gradients in a tar oil-contaminated aquifer. Appl. Geochem. 23:1715-1730.
-
- Bailly, J., L. Fraissinet-Tachet, J.-C. Verner, J.-C. Debaud, M. Lemaire, M. Wesolowski-Louvel, and R. Marmeisse. 2007. Soil eukaryotic functional diversity, a metatranscriptomic approach. ISME J. 1:632-642. - PubMed
-
- Bakermans, C., and E. L. Madsen. 2002. Detection in coal tar waste-contaminated groundwater of mRNA transcripts related to naphthalene dioxygenase by fluorescent in situ hybridization with tyramide signal amplification. J. Microbiol. Methods 50:75-84. - PubMed
-
- Bakermans, C., and E. L. Madsen. 2002. Diversity of 16S rDNA and naphthalene dioxygenase genes from coal-tar-waste-contaminated aquifer waters. Microb. Ecol. 44:95-106. - PubMed
-
- Bakermans, C., A. M. Hohnstock-Ashe, S. Padmanabhan, P. Padmanabhan, and E. L. Madsen. 2002. Geochemical and physiological evidence for mixed aerobic and anaerobic field biodegradation of coal tar waste by subsurface microbial communities. Microb. Ecol. 44:107-117. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

