Structure, diversity, and mobility of the Salmonella pathogenicity island 7 family of integrative and conjugative elements within Enterobacteriaceae
- PMID: 22247511
- PMCID: PMC3294861
- DOI: 10.1128/JB.06403-11
Structure, diversity, and mobility of the Salmonella pathogenicity island 7 family of integrative and conjugative elements within Enterobacteriaceae
Abstract
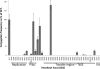
Integrative and conjugative elements (ICEs) are self-mobile genetic elements found in the genomes of some bacteria. These elements may confer a fitness advantage upon their host bacteria through the cargo genes that they carry. Salmonella pathogenicity island 7 (SPI-7), found within some pathogenic strains of Salmonella enterica, possesses features indicative of an ICE and carries genes implicated in virulence. We aimed to identify and fully analyze ICEs related to SPI-7 within the genus Salmonella and other Enterobacteriaceae. We report the sequence of two novel SPI-7-like elements, found within strains of Salmonella bongori, which share 97% nucleotide identity over conserved regions with SPI-7 and with each other. Although SPI-7 within Salmonella enterica serovar Typhi appears to be fixed within the chromosome, we present evidence that these novel elements are capable of excision and self-mobility. Phylogenetic analyses show that these Salmonella mobile elements share an ancestor which existed approximately 3.6 to 15.8 million years ago. Additionally, we identified more distantly related ICEs, with distinct cargo regions, within other strains of Salmonella as well as within Citrobacter, Erwinia, Escherichia, Photorhabdus, and Yersinia species. In total, we report on a collection of 17 SPI-7 related ICEs within enterobacterial species, of which six are novel. Using comparative and mutational studies, we have defined a core of 27 genes essential for conjugation. We present a growing family of SPI-7-related ICEs whose mobility, abundance, and cargo variability indicate that these elements may have had a large impact on the evolution of the Enterobacteriaceae.
Figures
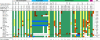


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

