Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq
- PMID: 18725932
- PMCID: PMC2515195
- DOI: 10.1371/journal.pgen.1000163
Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq
Abstract
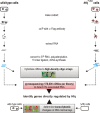
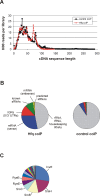
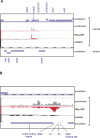
Recent advances in high-throughput pyrosequencing (HTPS) technology now allow a thorough analysis of RNA bound to cellular proteins, and, therefore, of post-transcriptional regulons. We used HTPS to discover the Salmonella RNAs that are targeted by the common bacterial Sm-like protein, Hfq. Initial transcriptomic analysis revealed that Hfq controls the expression of almost a fifth of all Salmonella genes, including several horizontally acquired pathogenicity islands (SPI-1, -2, -4, -5), two sigma factor regulons, and the flagellar gene cascade. Subsequent HTPS analysis of 350,000 cDNAs, derived from RNA co-immunoprecipitation (coIP) with epitope-tagged Hfq or control coIP, identified 727 mRNAs that are Hfq-bound in vivo. The cDNA analysis discovered new, small noncoding RNAs (sRNAs) and more than doubled the number of sRNAs known to be expressed in Salmonella to 64; about half of these are associated with Hfq. Our analysis explained aspects of the pleiotropic effects of Hfq loss-of-function. Specifically, we found that the mRNAs of hilD (master regulator of the SPI-1 invasion genes) and flhDC (flagellar master regulator) were bound by Hfq. We predicted that defective SPI-1 secretion and flagellar phenotypes of the hfq mutant would be rescued by overexpression of HilD and FlhDC, and we proved this to be correct. The combination of epitope-tagging and HTPS of immunoprecipitated RNA detected the expression of many intergenic chromosomal regions of Salmonella. Our approach overcomes the limited availability of high-density microarrays that have impeded expression-based sRNA discovery in microorganisms. We present a generic strategy that is ideal for the systems-level analysis of the post-transcriptional regulons of RNA-binding proteins and for sRNA discovery in a wide range of bacteria.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





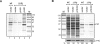
References
-
- Majdalani N, Vanderpool CK, Gottesman S. Bacterial small RNA regulators. Crit Rev Biochem Mol Biol. 2005;40:93–113. - PubMed
-
- Vogel J, Wagner EG. Target identification of regulatory sRNAs in bacteria. Curr Opin Microbiol. 2007;10:262–270. - PubMed
-
- Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu Rev Biochem. 2005;74:199–217. - PubMed
-
- Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol. 2007;10:134–139. - PubMed
-
- Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

