BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals
- PMID: 18391959
- PMCID: PMC2871289
- DOI: 10.1038/nmeth.1199
BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals
Erratum in
- Nat Methods. 2008 Aug;5(8):748
Abstract
The interpretation of genome sequences requires reliable and standardized methods to assess protein function at high throughput. Here we describe a fast and reliable pipeline to study protein function in mammalian cells based on protein tagging in bacterial artificial chromosomes (BACs). The large size of the BAC transgenes ensures the presence of most, if not all, regulatory elements and results in expression that closely matches that of the endogenous gene. We show that BAC transgenes can be rapidly and reliably generated using 96-well-format recombineering. After stable transfection of these transgenes into human tissue culture cells or mouse embryonic stem cells, the localization, protein-protein and/or protein-DNA interactions of the tagged protein are studied using generic, tag-based assays. The same high-throughput approach will be generally applicable to other model systems.
Figures
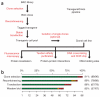
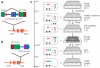
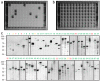
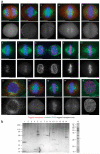

Comment in
-
BAC to the future: functional genomics in mammals.Nat Methods. 2008 May;5(5):383-4. doi: 10.1038/nmeth0508-383. Nat Methods. 2008. PMID: 18446155 Free PMC article.
-
For every protein its tag.Nat Methods. 2012 Oct;9(10):941. doi: 10.1038/nmeth.2187. Nat Methods. 2012. PMID: 23193563 No abstract available.
References
-
- Kittler R, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat. Cell Biol. 2007;9:1401–1412. - PubMed
-
- Paddison PJ, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. - PubMed
-
- Kittler R, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. - PubMed
-
- Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. - PubMed
-
- Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

