Evaluating the transcriptional regulators of arterial gene expression via a catalogue of characterized arterial enhancers
- PMID: 39819837
- PMCID: PMC11896612
- DOI: 10.7554/eLife.102440
Evaluating the transcriptional regulators of arterial gene expression via a catalogue of characterized arterial enhancers
Abstract
The establishment and growth of the arterial endothelium require the coordinated expression of numerous genes. However, regulation of this process is not yet fully understood. Here, we combined in silico analysis with transgenic mice and zebrafish models to characterize arterial-specific enhancers associated with eight key arterial identity genes (Acvrl1/Alk1, Cxcr4, Cxcl12, Efnb2, Gja4/Cx37, Gja5/Cx40, Nrp1, and Unc5b). Next, to elucidate the regulatory pathways upstream of arterial gene transcription, we investigated the transcription factors binding each arterial enhancer compared to a similar assessment of non-arterial endothelial enhancers. These results found that binding of SOXF and ETS factors was a common occurrence at both arterial and pan-endothelial enhancers, suggesting neither are sufficient to direct arterial specificity. Conversely, FOX motifs independent of ETS motifs were over-represented at arterial enhancers. Further, MEF2 and RBPJ binding was enriched but not ubiquitous at arterial enhancers, potentially linked to specific patterns of behaviour within the arterial endothelium. Lastly, there was no shared or arterial-specific signature for WNT-associated TCF/LEF, TGFβ/BMP-associated SMAD1/5 and SMAD2/3, shear stress-associated KLF4, or venous-enriched NR2F2. This cohort of well-characterized and in vivo-verified enhancers can now provide a platform for future studies into the interaction of different transcriptional and signaling pathways with arterial gene expression.
Keywords: SOXF transcription factors; arterial enhancer; arterial gene transcription; arteriovenous differentiation; developmental biology; genetics; genomics; mouse; transcriptional regulation; vascular development; zebrafish.
Plain language summary
Our blood vessels are a biological transport system that carry oxygen and nutrients to all the cells and tissues of our bodies. Each type of blood vessel has a different structure depending on its role. For example, arteries are large, strong-walled vessels that carry oxygenated blood away from the heart into the rest of the body, while veins carry blood back to the heart and lungs once all the oxygen has been used up. All blood vessels contain an inner lining made up of cells termed endothelial cells. These cells are also important for the formation of new blood vessels, which happens via a process called angiogenesis. During angiogenesis, the endothelial lining of new vessels forms first, by ‘sprouting’ or ‘splitting’ from the endothelial cells lining existing vessels. We know that angiogenesis is accompanied by changes in gene activity within the new endothelial cells. For example, during the development of new arteries, endothelial cells will turn on genes involved in artery formation. These changes are controlled by biological switches, which involve special proteins (called transcription factors) and DNA sequences close to specific genes (called enhancers). When the right transcription factor interacts with an enhancer for a gene, the gene ‘switches on’. Despite this, however, very few enhancers associated with arterial angiogenesis are known, and the mechanisms controlling this process are still poorly understood. Nornes et al. therefore set out to identify more arterial enhancers and study how they worked. To identify potential enhancers, Nornes et al. first used computer-based analysis of the DNA surrounding eight genes known to be involved in artery formation. The enhancers were then tested in zebrafish and mice to confirm their ability to switch genes on in artery endothelial cells. These experiments revealed a set of 15 new arterial enhancers, which were tested in further biochemical and genetic studies to determine which transcription factors could interact with them. Several transcription factors previously thought to be involved in artery development did not appear to interact with any of the new enhancers. This study sheds new light on the genetic control of blood vessel formation, in particular artery development. Nornes et al. hope that in the future the knowledge gained from these experiments will contribute to a better understanding of angiogenesis during early life, in health and disease.
© 2025, Nornes et al.
Conflict of interest statement
SN, SB, NA, IM, SD No competing interests declared
Figures

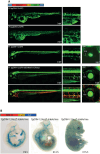

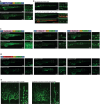



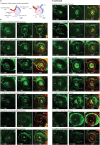

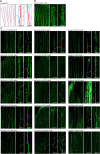












Update of
- doi: 10.1101/2024.04.30.591717
Comment in
- doi: 10.7554/eLife.106133
References
-
- Akerberg BN, Gu F, VanDusen NJ, Zhang X, Dong R, Li K, Zhang B, Zhou B, Sethi I, Ma Q, Wasson L, Wen T, Liu J, Dong K, Conlon FL, Zhou J, Yuan G-C, Zhou P, Pu WT. A reference map of murine cardiac transcription factor chromatin occupancy identifies dynamic and conserved enhancers. Nature Communications. 2019;10:e4907. doi: 10.1038/s41467-019-12812-3. - DOI - PMC - PubMed
-
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jørgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Müller F, Forrest ARR, Carninci P, Rehli M, Sandelin A, The FANTOM Consortium An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. - DOI - PMC - PubMed
-
- Andrade J, Shi C, Costa ASH, Choi J, Kim J, Doddaballapur A, Sugino T, Ong YT, Castro M, Zimmermann B, Kaulich M, Guenther S, Wilhelm K, Kubota Y, Braun T, Koh GY, Grosso AR, Frezza C, Potente M. Control of endothelial quiescence by FOXO-regulated metabolites. Nature Cell Biology. 2021;23:413–423. doi: 10.1038/s41556-021-00637-6. - DOI - PMC - PubMed
-
- Antonio M. VennRanges. 0.1Github. 2019 https://github.com/antonio-mora/vennRanges
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

