Genome-wide analysis reveals Sall4 to be a major regulator of pluripotency in murine-embryonic stem cells
- PMID: 19060217
- PMCID: PMC2604985
- DOI: 10.1073/pnas.0809321105
Genome-wide analysis reveals Sall4 to be a major regulator of pluripotency in murine-embryonic stem cells
Abstract
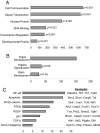
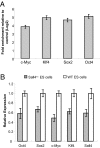
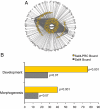
Embryonic stem cells have potential utility in regenerative medicine because of their pluripotent characteristics. Sall4, a zinc-finger transcription factor, is expressed very early in embryonic development with Oct4 and Nanog, two well-characterized pluripotency regulators. Sall4 plays an important role in governing the fate of stem cells through transcriptional regulation of both Oct4 and Nanog. By using chromatin immunoprecipitation coupled to microarray hybridization (ChIP-on-chip), we have mapped global gene targets of Sall4 to further investigate regulatory processes in W4 mouse ES cells. A total of 3,223 genes were identified that were bound by the Sall4 protein on duplicate assays with high confidence, and many of these have major functions in developmental and regulatory pathways. Sall4 bound approximately twice as many annotated genes within promoter regions as Nanog and approximately four times as many as Oct4. Immunoprecipitation revealed a heteromeric protein complex(es) between Sall4, Oct4, and Nanog, consistent with binding site co-occupancies. Decreasing Sall4 expression in W4 ES cells decreases the expression levels of Oct4, Sox2, c-Myc, and Klf4, four proteins capable of reprogramming somatic cells to an induced pluripotent state. Further, Sall4 bound many genes that are regulated in part by chromatin-based epigenetic events mediated by polycomb-repressive complexes and bivalent domains. This suggests that Sall4 plays a diverse role in regulating stem cell pluripotency during early embryonic development through integration of transcriptional and epigenetic controls.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Kohlhase J, et al. Isolation, characterization and organ-specific expression of two novel human zinc finger genes related to the Drosophila gene spalt. Genomics. 1996;38:291–298. - PubMed
-
- Kohlhase J, et al. SALL3, a new member of the human spalt-like gene family, maps to 18q23. Genomics. 1999;62:216–222. - PubMed
-
- Kuhnlein RP, Schuh R. Dual function of the region-specific homeotic gene spalt during Drosophila tracheal system development. Development. 1996;122:2215–2223. - PubMed
-
- Kohlhase J, et al. Okihiro syndrome is caused by SALL4 mutations. Hum Mol Genet. 2002;11:2979–2987. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

