An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans
- PMID: 18662544
- PMCID: PMC4719053
- DOI: 10.1016/j.cell.2008.05.044
An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans
Abstract
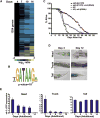
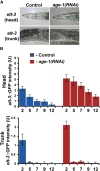
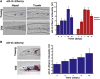
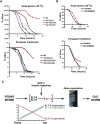
To define the C. elegans aging process at the molecular level, we used DNA microarray experiments to identify a set of 1294 age-regulated genes and found that the GATA transcription factors ELT-3, ELT-5, and ELT-6 are responsible for age regulation of a large fraction of these genes. Expression of elt-5 and elt-6 increases during normal aging, and both of these GATA factors repress expression of elt-3, which shows a corresponding decrease in expression in old worms. elt-3 regulates a large number of downstream genes that change expression in old age, including ugt-9, col-144, and sod-3. elt-5(RNAi) and elt-6(RNAi) worms have extended longevity, indicating that elt-3, elt-5, and elt-6 play an important functional role in the aging process. These results identify a transcriptional circuit that guides the rapid aging process in C. elegans and indicate that this circuit is driven by drift of developmental pathways rather than accumulation of damage.
Figures


Comment in
-
Re-evaluating the role of ELT-3 in a GATA transcription factor circuit proposed to guide aging in C. elegans.Mech Ageing Dev. 2012 Jan;133(1):50-3. doi: 10.1016/j.mad.2011.09.006. Epub 2011 Oct 6. Mech Ageing Dev. 2012. PMID: 22001047
References
-
- Anson RM, Hansford RG. Mitochondrial influence on aging rate in Caenorhabditis elegans. Aging Cell. 2004;3:29–34. - PubMed
-
- Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. - PubMed
-
- Aviv A. Telomeres and human aging: facts and fibs. Sci Aging Knowledge Environ. 2004;2004:pe43. - PubMed
-
- Ayub K, Hallett MB. Signalling shutdown strategies in aging immune cells. Aging Cell. 2004;3:145–149. - PubMed
-
- Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8:299–309. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

