Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells
- PMID: 19523850
- PMCID: PMC2709841
- DOI: 10.1016/j.immuni.2009.05.006
Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells
Abstract
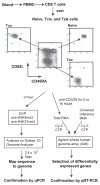
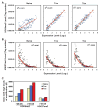
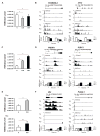
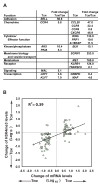
Memory lymphocytes are characterized by their ability to exhibit a rapid response to the recall antigen, in which differential transcription is important, yet the underlying mechanism is not understood. We report here a genome-wide analysis of histone methylation on two histone H3 lysine residues (H3K4me3 and H3K27me3) and gene expression profiles in naive and memory CD8(+) T cells. We found that specific correlation exists between gene expression and the amounts of H3K4me3 (positive correlation) and H3K27me3 (negative correlation) across the gene body. These correlations displayed four distinct modes (repressive, active, poised, and bivalent), reflecting different functions of these genes in CD8(+) T cells. Furthermore, a permissive chromatin state of each gene was established by a combination of different histone modifications. Our findings reveal a complex regulation by histone methylation in differential gene expression and suggest that histone methylation may be responsible for memory CD8(+) T cell function.
Figures



References
-
- Antia R, Ganusov VV, Ahmed R. The role of models in understanding CD8+ T-cell memory. Nat Rev Immunol. 2005;5:101–111. - PubMed
-
- Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, Kundu TK. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. 2004;279:51163–51171. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

