A CD4+ T cell reference map delineates subtype-specific adaptation during acute and chronic viral infections
- PMID: 35829695
- PMCID: PMC9323004
- DOI: 10.7554/eLife.76339
A CD4+ T cell reference map delineates subtype-specific adaptation during acute and chronic viral infections
Abstract
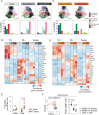
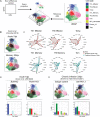
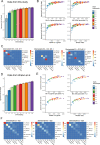
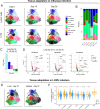
CD4+ T cells are critical orchestrators of immune responses against a large variety of pathogens, including viruses. While multiple CD4+ T cell subtypes and their key transcriptional regulators have been identified, there is a lack of consistent definition for CD4+ T cell transcriptional states. In addition, the progressive changes affecting CD4+ T cell subtypes during and after immune responses remain poorly defined. Using single-cell transcriptomics, we characterized the diversity of CD4+ T cells responding to self-resolving and chronic viral infections in mice. We built a comprehensive map of virus-specific CD4+ T cells and their evolution over time, and identified six major cell states consistently observed in acute and chronic infections. During the course of acute infections, T cell composition progressively changed from effector to memory states, with subtype-specific gene modules and kinetics. Conversely, in persistent infections T cells acquired distinct, chronicity-associated programs. By single-cell T cell receptor (TCR) analysis, we characterized the clonal structure of virus-specific CD4+ T cells across individuals. Virus-specific CD4+ T cell responses were essentially private across individuals and most T cells differentiated into both Tfh and Th1 subtypes irrespective of their TCR. Finally, we showed that our CD4+ T cell map can be used as a reference to accurately interpret cell states in external single-cell datasets across tissues and disease models. Overall, this study describes a previously unappreciated level of adaptation of the transcriptional states of CD4+ T cells responding to viruses and provides a new computational resource for CD4+ T cell analysis.
Keywords: CD4 T cells; exhaustion; gene expression; immunology; inflammation; mouse; single-cell data science; single-cell transcriptomics; viral infection.
Conflict of interest statement
MA, AT, ZS, MK, TC, SC No competing interests declared
Figures











References
-
- Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice Role in suppression of cytotoxic T lymphocyte response and viral persistence. The Journal of Experimental Medicine. 1984;160:521–540. doi: 10.1084/jem.160.2.521. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- figshare/10.6084/m9.figshare.16592693
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

