Deciphering the combinatorial landscape of immunity
- PMID: 33225996
- PMCID: PMC7748411
- DOI: 10.7554/eLife.62148
Deciphering the combinatorial landscape of immunity
Abstract
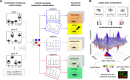
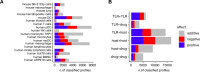
From cellular activation to drug combinations, immunological responses are shaped by the action of multiple stimuli. Synergistic and antagonistic interactions between stimuli play major roles in shaping immune processes. To understand combinatorial regulation, we present the immune Synergistic/Antagonistic Interaction Learner (iSAIL). iSAIL includes a machine learning classifier to map and interpret interactions, a curated compendium of immunological combination treatment datasets, and their global integration into a landscape of ~30,000 interactions. The landscape is mined to reveal combinatorial control of interleukins, checkpoints, and other immune modulators. The resource helps elucidate the modulation of a stimulus by interactions with other cofactors, showing that TNF has strikingly different effects depending on co-stimulators. We discover new functional synergies between TNF and IFNβ controlling dendritic cell-T cell crosstalk. Analysis of laboratory or public combination treatment studies with this user-friendly web-based resource will help resolve the complex role of interaction effects on immune processes.
Keywords: combination treatment experiment; computational biology; cytokine interactions; dendritic cell T cell cross-talk; human; immunology; inflammation; machine learning; mouse; signal integration; synergy antagonism; systems biology.
© 2020, Cappuccio et al.
Conflict of interest statement
AC, SJ, BH, SS, VS, EZ No competing interests declared
Figures


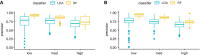



References
-
- Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, Berger P, Desborough V, Smith T, Campbell J, Thomson E, Monteiro R, Guimaraes P, Walters B, Wiser J, Butte AJ. ImmPort: disseminating data to the public for the future of immunology. Immunologic Research. 2014;58:234–239. doi: 10.1007/s12026-014-8516-1. - DOI - PubMed
-
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G. A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nature Cell Biology. 2004;6:97–105. doi: 10.1038/ncb1086. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- IT-DC 281987/ERC_/European Research Council/International
- ANR-11-LABX-0043 CIC IGR-Curie 1428/Agence Nationale de la Recherche/International
- U19 AI117873/AI/NIAID NIH HHS/United States
- 680890/ERC_/European Research Council/International
- N6600119C4022/Defense Advanced Research Projects Agency/International
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

