Probe-Seq enables transcriptional profiling of specific cell types from heterogeneous tissue by RNA-based isolation
- PMID: 31815670
- PMCID: PMC6901332
- DOI: 10.7554/eLife.51452
Probe-Seq enables transcriptional profiling of specific cell types from heterogeneous tissue by RNA-based isolation
Abstract
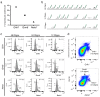
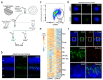
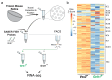
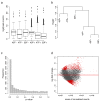
Recent transcriptional profiling technologies are uncovering previously-undefined cell populations and molecular markers at an unprecedented pace. While single cell RNA (scRNA) sequencing is an attractive approach for unbiased transcriptional profiling of all cell types, a complementary method to isolate and sequence specific cell populations from heterogeneous tissue remains challenging. Here, we developed Probe-Seq, which allows deep transcriptional profiling of specific cell types isolated using RNA as the defining feature. Dissociated cells are labeled using fluorescent in situ hybridization (FISH) for RNA, and then isolated by fluorescent activated cell sorting (FACS). We used Probe-Seq to purify and profile specific cell types from mouse, human, and chick retinas, as well as from Drosophila midguts. Probe-Seq is compatible with frozen nuclei, making cell types within archival tissue immediately accessible. As it can be multiplexed, combinations of markers can be used to create specificity. Multiplexing also allows for the isolation of multiple cell types from one cell preparation. Probe-Seq should enable RNA profiling of specific cell types from any organism.
Keywords: D. melanogaster; FISH; Probe-Seq; RNA-sequencing; cell types; chicken; genetics; genomics; human; mouse; neuroscience; retina; transcriptome.
© 2019, Amamoto et al.
Conflict of interest statement
RA, MG, EW, JC, SL, EL, NP, CC No competing interests declared
Figures

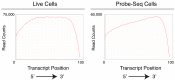
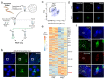
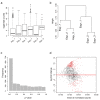
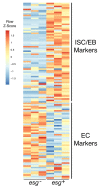
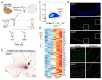
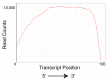
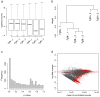
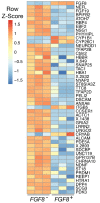
Similar articles
-
Probe-Seq: Method for RNA Sequencing of Specific Cell Types from Animal Tissue.Bio Protoc. 2020 Sep 20;10(18):e3749. doi: 10.21769/BioProtoc.3749. eCollection 2020 Sep 20. Bio Protoc. 2020. PMID: 33659409 Free PMC article.
-
NKX2-2 based nuclei sorting on frozen human archival pancreas enables the enrichment of islet endocrine populations for single-nucleus RNA sequencing.BMC Genomics. 2024 Apr 30;25(1):427. doi: 10.1186/s12864-024-10335-w. BMC Genomics. 2024. PMID: 38689254 Free PMC article.
-
Hydrop enables droplet-based single-cell ATAC-seq and single-cell RNA-seq using dissolvable hydrogel beads.Elife. 2022 Feb 23;11:e73971. doi: 10.7554/eLife.73971. Elife. 2022. PMID: 35195064 Free PMC article.
-
Single-cell RNA-sequencing of retrieved human oocytes and eggs in clinical practice and for human ovarian cell atlasing.Mol Reprod Dev. 2022 Dec;89(12):597-607. doi: 10.1002/mrd.23648. Epub 2022 Oct 20. Mol Reprod Dev. 2022. PMID: 36264989 Free PMC article. Review.
-
RNA-Seq methods for transcriptome analysis.Wiley Interdiscip Rev RNA. 2017 Jan;8(1):10.1002/wrna.1364. doi: 10.1002/wrna.1364. Epub 2016 May 19. Wiley Interdiscip Rev RNA. 2017. PMID: 27198714 Free PMC article. Review.
Cited by
-
Methods and tools for spatial mapping of single-cell RNAseq clusters in Drosophila.Genetics. 2021 Apr 15;217(4):iyab019. doi: 10.1093/genetics/iyab019. Genetics. 2021. PMID: 33713129 Free PMC article. Review.
-
High-throughput single cell -omics using semi-permeable capsules.bioRxiv [Preprint]. 2025 Mar 17:2025.03.14.642805. doi: 10.1101/2025.03.14.642805. bioRxiv. 2025. PMID: 40166174 Free PMC article. Preprint.
-
Unveiling heterogeneity in rare cells by combining RNA-based sorting and scRNA-seq.Nat Genet. 2025 Feb;57(2):283-284. doi: 10.1038/s41588-024-02073-2. Nat Genet. 2025. PMID: 39920268 No abstract available.
-
Transcript-specific enrichment enables profiling of rare cell states via single-cell RNA sequencing.Nat Genet. 2025 Feb;57(2):451-460. doi: 10.1038/s41588-024-02036-7. Epub 2025 Jan 8. Nat Genet. 2025. PMID: 39779958 Free PMC article.
-
Transcript-specific enrichment enables profiling rare cell states via scRNA-seq.bioRxiv [Preprint]. 2024 Mar 27:2024.03.27.587039. doi: 10.1101/2024.03.27.587039. bioRxiv. 2024. Update in: Nat Genet. 2025 Feb;57(2):451-460. doi: 10.1038/s41588-024-02036-7. PMID: 38586040 Free PMC article. Updated. Preprint.
References
-
- Aken BL, Achuthan P, Akanni W, Amode MR, Bernsdorff F, Bhai J, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Gil L, Girón CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Juettemann T, Keenan S, Laird MR, Lavidas I, Maurel T, McLaren W, Moore B, Murphy DN, Nag R, Newman V, Nuhn M, Ong CK, Parker A, Patricio M, Riat HS, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Wilder SP, Zadissa A, Kostadima M, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Cunningham F, Yates A, Zerbino DR, Flicek P. Ensembl 2017. Nucleic Acids Research. 2017;45:D635–D642. doi: 10.1093/nar/gkw1104. - DOI - PMC - PubMed
-
- Alles J, Karaiskos N, Praktiknjo SD, Grosswendt S, Wahle P, Ruffault PL, Ayoub S, Schreyer L, Boltengagen A, Birchmeier C, Zinzen R, Kocks C, Rajewsky N. Cell fixation and preservation for droplet-based single-cell transcriptomics. BMC Biology. 2017;15:44. doi: 10.1186/s12915-017-0383-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

