The noncanonical RNA-binding protein RAN stabilizes the mRNA of intranuclear stress granule assembly factor G3BP1 in nasopharyngeal carcinoma
- PMID: 39510185
- PMCID: PMC11635782
- DOI: 10.1016/j.jbc.2024.107964
The noncanonical RNA-binding protein RAN stabilizes the mRNA of intranuclear stress granule assembly factor G3BP1 in nasopharyngeal carcinoma
Abstract
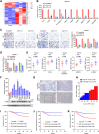
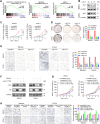
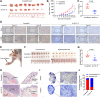
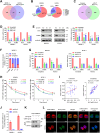
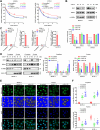
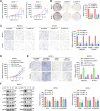
RNA-binding proteins (RBPs) play critical roles in tumor progression by participating in the posttranscriptional regulation of RNA. However, the levels and function of RBPs in nasopharyngeal carcinoma (NPC) remain elusive. Here we identified a noncanonical RBP RAN that has the most significant role in NPC progression by a small siRNA pool screening. Functionally, RAN facilitates NPC proliferation and metastasis in vitro and in vivo. High levels of RAN are associated with poor prognosis of NPC patients and can be performed as a prognostic biomarker. Mechanistically, RAN increases the nucleus import of TDP43 and enhances TDP43 nuclear distribution. On the other hand, RAN is directly bound to the coding sequence of G3BP1 mRNA and serves as an adapter to facilitate TDP43 interacting with G3BP1 mRNA 3' UTR. These contribute to increasing G3BP1 mRNA stability in the nucleus and lead to upregulation of G3BP1, which further enhances AKT and ERK signaling and ultimately promotes NPC proliferation and metastasis. These findings reveal that RAN stabilizes intranuclear G3BP1 mRNA by dual mechanisms: recruiting TDP43 into the nucleus and enhancing its interaction with G3BP1 mRNA, suggesting a critical role of RAN in NPC progression and providing a new regulation framework of RBP-RNA.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Zhao Y., Mir C., Garcia-Mayea Y., Paciucci R., Kondoh H., LLeonart M.E. RNA-binding proteins: underestimated contributors in tumorigenesis. Semin. Cancer Biol. 2022;86:431–444. - PubMed
-
- Gebauer F., Schwarzl T., Valcárcel J., Hentze M.W. RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 2021;22:185–198. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

