FMRP promotes RNA localization to neuronal projections through interactions between its RGG domain and G-quadruplex RNA sequences
- PMID: 32510328
- PMCID: PMC7279889
- DOI: 10.7554/eLife.52621
FMRP promotes RNA localization to neuronal projections through interactions between its RGG domain and G-quadruplex RNA sequences
Abstract
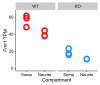
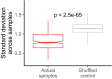
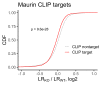
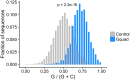
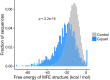
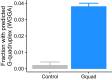
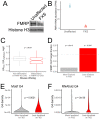
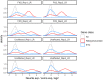
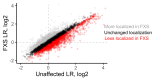
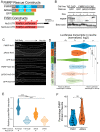
The sorting of RNA molecules to subcellular locations facilitates the activity of spatially restricted processes. We have analyzed subcellular transcriptomes of FMRP-null mouse neuronal cells to identify transcripts that depend on FMRP for efficient transport to neurites. We found that these transcripts contain an enrichment of G-quadruplex sequences in their 3' UTRs, suggesting that FMRP recognizes them to promote RNA localization. We observed similar results in neurons derived from Fragile X Syndrome patients. We identified the RGG domain of FMRP as important for binding G-quadruplexes and the transport of G-quadruplex-containing transcripts. Finally, we found that the translation and localization targets of FMRP were distinct and that an FMRP mutant that is unable to bind ribosomes still promoted localization of G-quadruplex-containing messages. This suggests that these two regulatory modes of FMRP may be functionally separated. These results provide a framework for the elucidation of similar mechanisms governed by other RNA-binding proteins.
Keywords: FMRP; RNA localization; chromosomes; fragile x syndrome; gene expression; genetics; genomics; human; mouse.
© 2020, Goering et al.
Conflict of interest statement
RG, LH, BG, NR, GB, HR, DD, JT No competing interests declared
Figures







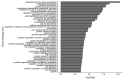



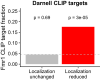
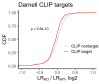
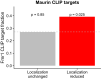
















References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

