Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells
- PMID: 19303854
- PMCID: PMC2716120
- DOI: 10.1016/j.cell.2008.12.043
Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells
Abstract
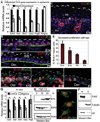
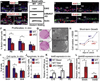
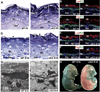
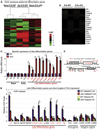
Although in vitro studies of embryonic stem cells have identified polycomb repressor complexes (PRCs) as key regulators of differentiation, it remains unclear as to how PRC-mediated mechanisms control fates of multipotent progenitors in developing tissues. Here, we show that an essential PRC component, Ezh2, is expressed in epidermal progenitors but diminishes concomitant with embryonic differentiation and with postnatal decline in proliferative activity. We show that Ezh2 controls proliferative potential of basal progenitors by repressing the Ink4A-Ink4B locus and tempers the developmental rate of differentiation by preventing premature recruitment of AP1 transcriptional activator to the structural genes that are required for epidermal differentiation. Together, our studies reveal that PRCs control epigenetic modifications temporally and spatially in tissue-restricted stem cells. They maintain their proliferative potential and globally repressing undesirable differentiation programs while selectively establishing a specific terminal differentiation program in a stepwise fashion.
Figures


Comment in
-
Polycomb repression under the skin.Cell. 2009 Mar 20;136(6):992-4. doi: 10.1016/j.cell.2009.03.004. Cell. 2009. PMID: 19303840
References
-
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
-
- Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. - PubMed
-
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

