EKLF/KLF1 expression defines a unique macrophage subset during mouse erythropoiesis
- PMID: 33570494
- PMCID: PMC7932694
- DOI: 10.7554/eLife.61070
EKLF/KLF1 expression defines a unique macrophage subset during mouse erythropoiesis
Abstract
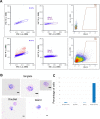
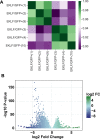
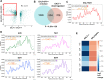
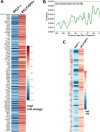
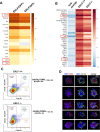
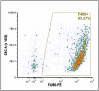
Erythroblastic islands are a specialized niche that contain a central macrophage surrounded by erythroid cells at various stages of maturation. However, identifying the precise genetic and transcriptional control mechanisms in the island macrophage remains difficult due to macrophage heterogeneity. Using unbiased global sequencing and directed genetic approaches focused on early mammalian development, we find that fetal liver macrophages exhibit a unique expression signature that differentiates them from erythroid and adult macrophage cells. The importance of erythroid Krüppel-like factor (EKLF)/KLF1 in this identity is shown by expression analyses in EKLF-/- and in EKLF-marked macrophage cells. Single-cell sequence analysis simplifies heterogeneity and identifies clusters of genes important for EKLF-dependent macrophage function and novel cell surface biomarkers. Remarkably, this singular set of macrophage island cells appears transiently during embryogenesis. Together, these studies provide a detailed perspective on the importance of EKLF in the establishment of the dynamic gene expression network within erythroblastic islands in the developing embryo and provide the means for their efficient isolation.
Keywords: EKLF/Klf1; developmental biology; erythropoiesis; fetal liver island macrophage; mouse; single cell sequencing; transcription factors.
© 2021, Mukherjee et al.
Conflict of interest statement
KM, LX, AP, MG, AC, JB No competing interests declared
Figures










References
-
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder--open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. - DOI - PMC - PubMed
-
- Buesche G, Teoman H, Giagounidis A, Göhring G, Schlegelberger B, Ganser A, Aul C, Kreipe HH. Impaired formation of erythroblastic islands is associated with erythroid failure and poor prognosis in a significant proportion of patients with myelodysplastic syndromes. Haematologica. 2016;101:e177–e181. doi: 10.3324/haematol.2015.129015. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

