Exploiting the full power of temporal gene expression profiling through a new statistical test: application to the analysis of muscular dystrophy data
- PMID: 16584545
- PMCID: PMC1450310
- DOI: 10.1186/1471-2105-7-183
Exploiting the full power of temporal gene expression profiling through a new statistical test: application to the analysis of muscular dystrophy data
Abstract
Background: The identification of biologically interesting genes in a temporal expression profiling dataset is challenging and complicated by high levels of experimental noise. Most statistical methods used in the literature do not fully exploit the temporal ordering in the dataset and are not suited to the case where temporal profiles are measured for a number of different biological conditions. We present a statistical test that makes explicit use of the temporal order in the data by fitting polynomial functions to the temporal profile of each gene and for each biological condition. A Hotelling T2-statistic is derived to detect the genes for which the parameters of these polynomials are significantly different from each other.
Results: We validate the temporal Hotelling T2-test on muscular gene expression data from four mouse strains which were profiled at different ages: dystrophin-, beta-sarcoglycan and gamma-sarcoglycan deficient mice, and wild-type mice. The first three are animal models for different muscular dystrophies. Extensive biological validation shows that the method is capable of finding genes with temporal profiles significantly different across the four strains, as well as identifying potential biomarkers for each form of the disease. The added value of the temporal test compared to an identical test which does not make use of temporal ordering is demonstrated via a simulation study, and through confirmation of the expression profiles from selected genes by quantitative PCR experiments. The proposed method maximises the detection of the biologically interesting genes, whilst minimising false detections.
Conclusion: The temporal Hotelling T2-test is capable of finding relatively small and robust sets of genes that display different temporal profiles between the conditions of interest. The test is simple, it can be used on gene expression data generated from any experimental design and for any number of conditions, and it allows fast interpretation of the temporal behaviour of genes. The R code is available from V.V. The microarray data have been submitted to GEO under series GSE1574 and GSE3523.
Figures

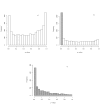
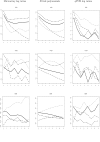
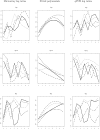
References
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

