GPRC5A promotes lung colonization of esophageal squamous cell carcinoma
- PMID: 39550386
- PMCID: PMC11569164
- DOI: 10.1038/s41467-024-54251-9
GPRC5A promotes lung colonization of esophageal squamous cell carcinoma
Abstract
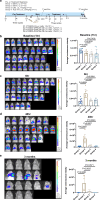
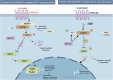
Emerging evidence suggests that cancer cells may disseminate early, prior to the formation of traditional macro-metastases. However, the mechanisms underlying the seeding and transition of early disseminated cancer cells (DCCs) into metastatic tumors remain poorly understood. Through single-cell RNA sequencing, we show that early lung DCCs from esophageal squamous cell carcinoma (ESCC) exhibit a trophoblast-like 'tumor implantation' phenotype, which enhances their dissemination and supports metastatic growth. Notably, ESCC cells overexpressing GPRC5A demonstrate improved implantation and persistence, resulting in macro-metastases in the lungs. Clinically, elevated GPRC5A level is associated with poorer outcomes in a cohort of 148 ESCC patients. Mechanistically, GPRC5A is found to potentially interact with WWP1, facilitating the polyubiquitination and degradation of LATS1, thereby activating YAP1 signaling pathways essential for metastasis. Importantly, targeting YAP1 axis with CA3 or TED-347 significantly diminishes early implantation and macro-metastases. Thus, the GPRC5A/WWP1/LATS1/YAP1 pathway represents a crucial target for therapeutic intervention in ESCC lung metastases.
© 2024. The Author(s).
Conflict of interest statement
Figures





References
-
- Klein, C. A. Cancer progression and the invisible phase of metastatic colonization. Nat Rev Cancer20, 681–694 (2020). - PubMed
-
- Pantel, K., Alix-Panabières, C. & Riethdorf, S. Cancer micrometastases. Nat Rev Clin Oncol6, 339–351 (2009). - PubMed
-
- Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J Clin74, 12–49 (2024). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

