Cell-specific gene expression in Langerhans cell histiocytosis lesions reveals a distinct profile compared with epidermal Langerhans cells
- PMID: 20220088
- PMCID: PMC3142675
- DOI: 10.4049/jimmunol.0902336
Cell-specific gene expression in Langerhans cell histiocytosis lesions reveals a distinct profile compared with epidermal Langerhans cells
Abstract
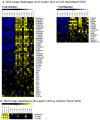
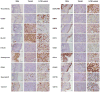
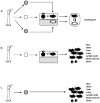
Langerhans cell histiocytosis (LCH) is a rare disease characterized by heterogeneous lesions containing CD207(+) Langerhans cells (LCs) and lymphocytes that can arise in almost any tissue and cause significant morbidity and mortality. After decades of research, the cause of LCH remains speculative. A prevailing model suggests that LCH arises from malignant transformation and metastasis of epidermal LCs. In this study, CD207(+) cells and CD3(+) T cells were isolated from LCH lesions to determine cell-specific gene expression. Compared with control epidermal CD207(+) cells, the LCH CD207(+) cells yielded 2113 differentially expressed genes (false discovery rate < 0.01). Surprisingly, the expression of many genes previously associated with LCH, including cell-cycle regulators, proinflammatory cytokines, and chemokines, were not significantly different from control LCs in our study. However, several novel genes whose products activate and recruit T cells to sites of inflammation, including SPP1 (osteopontin), were highly overexpressed in LCH CD207(+) cells. Furthermore, several genes associated with immature myeloid dendritic cells were overexpressed in LCH CD207(+) cells. Compared with the peripheral CD3(+) cells from LCH patients, the LCH lesion CD3(+) cells yielded only 162 differentially regulated genes (false discovery rate < 0.01), and the expression profile of the LCH lesion CD3(+) cells was consistent with an activated regulatory T cell phenotype with increased expression of FOXP3, CTLA4, and SPP1. Results from this study support a model of LCH pathogenesis in which lesions do not arise from epidermal LCs but from accumulation of bone marrow-derived immature myeloid dendritic cells that recruit activated lymphocytes.
Figures


References
-
- Carstensen H, Ornvold K. Langerhans-cell histiocytosis (histiocytosis X) in children. Ugeskr Laeger. 1993;155:1779–1783. - PubMed
-
- Arico M, Girschikofsky M, Genereau T, Klersy C, McClain K, Grois N, Emile JF, Lukina E, De JE, Danesino C. Langerhans cell histiocytosis in adults. Report from the International Registry of the Histiocyte Society. Eur J Cancer. 2003;39:2341–2348. - PubMed
-
- Arceci RJ. The histiocytoses: the fall of the Tower of Babel. Eur J Cancer. 1999;35:747–767. - PubMed
-
- McClain KL, Natkunam Y, Swerdlow SH. Atypical cellular disorders. Hematology Am Soc Hematol Educ Program. 2004:283–296. - PubMed
-
- Gadner H, Grois N, Potschger U, Minkov M, Arico M, Braier J, Broadbent V, Donadieu J, Henter JI, McCarter R, Ladisch S. Improved outcome in multisystem Langerhans cell histiocytosis is associated with therapy intensification. Blood. 2008;111:2556–2562. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

