m6A modification of U6 snRNA modulates usage of two major classes of pre-mRNA 5' splice site
- PMID: 36409063
- PMCID: PMC9803359
- DOI: 10.7554/eLife.78808
m6A modification of U6 snRNA modulates usage of two major classes of pre-mRNA 5' splice site
Abstract
Alternative splicing of messenger RNAs is associated with the evolution of developmentally complex eukaryotes. Splicing is mediated by the spliceosome, and docking of the pre-mRNA 5' splice site into the spliceosome active site depends upon pairing with the conserved ACAGA sequence of U6 snRNA. In some species, including humans, the central adenosine of the AC
Keywords: A. thaliana; C. elegans; D. melanogaster; ambient temperature; chromosomes; epitranscriptome; flowering time; gene expression; genetics; genomics; human; m6A; splicing; zebrafish.
Plain language summary
All the information necessary to build the proteins that perform the biological processes required for life is encoded in the DNA of an organism. Making these proteins requires the DNA sequence of a gene to be transcribed into a ‘messenger RNA’ (mRNA), which is then processed into a final, mature form. This blueprint is then translated to assemble the corresponding protein. When an mRNA is processed, segments of the sequence that do not code for protein are removed and the remaining coding sequences are joined together in the right order. An intricate molecular machine known as the spliceosome controls this mechanism by recognising the ‘splice sites’ where coding and non-coding sequences meet. Depending on external conditions, the spliceosome can ‘pick-and-mix’ the coding sequences to create different processed mRNAs (and therefore proteins) from a single gene. This alternative splicing mechanism is often used to regulate when certain biological processes take place based on environmental cues; for example, the splicing of genes which control the timing of plant flowering is sensitive to ambient temperatures. To investigate this mechanism, Parker et al. focused on Arabidopsis thaliana, a plant that blooms later when temperatures are low. This precise timing partly relies on a gene whose mRNA is efficiently spliced in the cold, resulting in an active form of its protein that blocks blooming. Parker et al. grew and screened many A. thaliana plants to find individuals that could flower early in the cold, in which splicing of this gene was disrupted. A mutant fitting these criteria was identified and subjected to further investigation, which revealed that it could not produce FIONA1. In non-mutant plants, this enzyme chemically modifies one of the components of the spliceosome, a small nuclear RNA known as U6. Parker et al found that there are two types of splice site – one more likely to interact with U6 and another that preferentially interacts with another small nuclear RNA, U5. When FIONA1 is inactive (such as in the mutant identified by Parker et al.), splice sites that tend to strongly interact with U5 are selected. However, when the enzyme is active, splice sites that tend to bind with the chemically modified U6 are used instead. Further work by Parker et al. showed that these two types of splice sites (‘preferring’ either U5 or U6) are found in equal proportions in the genomes of many species, including humans. This suggests that Parker et al. have uncovered an essential feature of how genomes are organised and splicing is controlled.
© 2022, Parker et al.
Conflict of interest statement
MP, BS, JK, AL, KK, NJ, FB, AS, GB, SF, BD, GS No competing interests declared
Figures








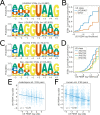
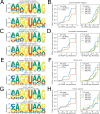
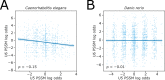
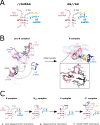


References
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR. Genome-Wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. - DOI - PubMed
-
- Andrews S. FastQC: A quality control tool for high throughput sequence data. v3Babraham Bioinformatics. 2017 https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- BB/M010066/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/T007222/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/W002302/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/W007967/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/V010662/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M004155/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 220212/Z/20/Z/WT_/Wellcome Trust/United Kingdom
- BB/W007673/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M000338/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

