Protective function and durability of mouse lymph node-resident memory CD8+ T cells
- PMID: 34143731
- PMCID: PMC8213409
- DOI: 10.7554/eLife.68662
Protective function and durability of mouse lymph node-resident memory CD8+ T cells
Abstract
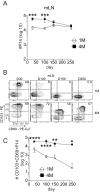
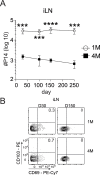
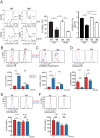
Protective lung tissue-resident memory CD8+T cells (Trm) form after influenza A virus (IAV) infection. We show that IAV infection of mice generates CD69+CD103+and other memory CD8+T cell populations in lung-draining mediastinal lymph nodes (mLNs) from circulating naive or memory CD8+T cells. Repeated antigen exposure, mimicking seasonal IAV infections, generates quaternary memory (4M) CD8+T cells that protect mLN from viral infection better than 1M CD8+T cells. Better protection by 4M CD8+T cells associates with enhanced granzyme A/B expression and stable maintenance of mLN CD69+CD103+4M CD8+T cells, vs the steady decline of CD69+CD103+1M CD8+T cells, paralleling the durability of protective CD69+CD103+4M vs 1M in the lung after IAV infection. Coordinated upregulation in canonical Trm-associated genes occurs in circulating 4M vs 1M populations without the enrichment of canonical downregulated Trm genes. Thus, repeated antigen exposure arms circulating memory CD8+T cells with enhanced capacity to form long-lived populations of Trm that enhance control of viral infections of the mLN.
Keywords: CD8+ T cell; CD8+ T cell resident memory; immunology; inflammation; lung; lymph node; mouse; repeated antigen exposure; virus infection.
© 2021, Anthony et al.
Conflict of interest statement
SA, NV, SM, Sv, QS, RV, RS, SH, IJ, SV, NB, HX, VB, JH No competing interests declared
Figures






References
-
- Beura LK, Wijeyesinghe S, Thompson EA, Macchietto MG, Rosato PC, Pierson MJ, Schenkel JM, Mitchell JS, Vezys V, Fife BT, Shen S, Masopust D. T Cells in Nonlymphoid Tissues Give Rise to Lymph-Node-Resident Memory T Cells. Immunity. 2018;48:e325–e338. doi: 10.1016/j.immuni.2018.01.015. - DOI - PMC - PubMed
-
- Cheuk S, Schlums H, Gallais Sérézal I, Martini E, Chiang SC, Marquardt N, Gibbs A, Detlofsson E, Introini A, Forkel M, Höög C, Tjernlund A, Michaëlsson J, Folkersen L, Mjösberg J, Blomqvist L, Ehrström M, Ståhle M, Bryceson YT, Eidsmo L. CD49a Expression Defines Tissue-Resident CD8 + T Cells Poised for Cytotoxic Function in Human Skin. Immunity. 2017;46:287–300. doi: 10.1016/j.immuni.2017.01.009. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R35 GM134880/GM/NIGMS NIH HHS/United States
- I01 BX002903/BX/BLRD VA/United States
- T32 AI007260/AI/NIAID NIH HHS/United States
- R01 AI124093/AI/NIAID NIH HHS/United States
- R01 AI139874/AI/NIAID NIH HHS/United States
- R01 AI042767/AI/NIAID NIH HHS/United States
- R01 AI127481/AI/NIAID NIH HHS/United States
- R01 AI125446/AI/NIAID NIH HHS/United States
- R01 AI114543/AI/NIAID NIH HHS/United States
- R37 AI042767/AI/NIAID NIH HHS/United States
- R01 AI121080/AI/NIAID NIH HHS/United States
- T32 AI007511/AI/NIAID NIH HHS/United States
- R21 AI042767/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

