Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts
- PMID: 19881528
- PMCID: PMC2958040
- DOI: 10.1038/ng.471
Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts
Abstract
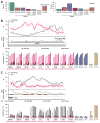
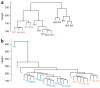
Induced pluripotent stem (iPS) cells are derived by epigenetic reprogramming, but their DNA methylation patterns have not yet been analyzed on a genome-wide scale. Here, we find substantial hypermethylation and hypomethylation of cytosine-phosphate-guanine (CpG) island shores in nine human iPS cell lines as compared to their parental fibroblasts. The differentially methylated regions (DMRs) in the reprogrammed cells (denoted R-DMRs) were significantly enriched in tissue-specific (T-DMRs; 2.6-fold, P < 10(-4)) and cancer-specific DMRs (C-DMRs; 3.6-fold, P < 10(-4)). Notably, even though the iPS cells are derived from fibroblasts, their R-DMRs can distinguish between normal brain, liver and spleen cells and between colon cancer and normal colon cells. Thus, many DMRs are broadly involved in tissue differentiation, epigenetic reprogramming and cancer. We observed colocalization of hypomethylated R-DMRs with hypermethylated C-DMRs and bivalent chromatin marks, and colocalization of hypermethylated R-DMRs with hypomethylated C-DMRs and the absence of bivalent marks, suggesting two mechanisms for epigenetic reprogramming in iPS cells and cancer.
Figures
References
-
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. - PubMed
-
- Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. - PubMed
-
- Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. - PubMed
-
- Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

