T-follicular helper cells are epigenetically poised to transdifferentiate into T-regulatory type 1 cells
- PMID: 39576679
- PMCID: PMC11584177
- DOI: 10.7554/eLife.97665
T-follicular helper cells are epigenetically poised to transdifferentiate into T-regulatory type 1 cells
Abstract
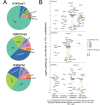
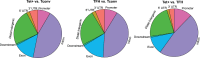
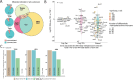
Chronic antigenic stimulation can trigger the formation of interleukin 10 (IL-10)-producing T-regulatory type 1 (TR1) cells in vivo. We have recently shown that murine T-follicular helper (TFH) cells are precursors of TR1 cells and that the TFH-to-TR1 cell transdifferentiation process is characterized by the progressive loss and acquisition of opposing transcription factor gene expression programs that evolve through at least one transitional cell stage. Here, we use a broad range of bulk and single-cell transcriptional and epigenetic tools to investigate the epigenetic underpinnings of this process. At the single-cell level, the TFH-to-TR1 cell transition is accompanied by both, downregulation of TFH cell-specific gene expression due to loss of chromatin accessibility, and upregulation of TR1 cell-specific genes linked to chromatin regions that remain accessible throughout the transdifferentiation process, with minimal generation of new open chromatin regions. By interrogating the epigenetic status of accessible TR1 genes on purified TFH and conventional T-cells, we find that most of these genes, including Il10, are already poised for expression at the TFH cell stage. Whereas these genes are closed and hypermethylated in Tconv cells, they are accessible, hypomethylated, and enriched for H3K27ac-marked and hypomethylated active enhancers in TFH cells. These enhancers are enriched for binding sites for the TFH and TR1-associated transcription factors TOX-2, IRF4, and c-MAF. Together, these data suggest that the TR1 gene expression program is genetically imprinted at the TFH cell stage.
Keywords: T-follicular helper cells; T-regulatory type 1 cells; TFH cells; TR1 cells; epigenetics; immunology; immunoregulation; inflammation; mouse; pMHCII-based nanomedicines; peptide-major histocompatibility complex class II - based nanomedicines; transdifferentiation.
© 2024, Garnica et al.
Conflict of interest statement
JG, PS, JY, JM, DM, CF, PS No competing interests declared, PS P. Santamaria is founder, scientific officer and stockholder of Parvus Therapeutics and receives funding from the company. He also has a consulting agreement with Sanofi
Figures











Update of
- doi: 10.1101/2024.05.17.594762
- doi: 10.7554/eLife.97665.1
- doi: 10.7554/eLife.97665.2
Comment in
- doi: 10.7554/elife.104239
References
-
- Araki Y, Wang Z, Zang C, Wood WH, III, Schones D, Cui K, Roh T-Y, Lhotsky B, Wersto RP, Peng W, Becker KG, Zhao K, Weng N. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity. 2009;30:912–925. doi: 10.1016/j.immuni.2009.05.006. - DOI - PMC - PubMed
-
- Belk JA, Yao W, Ly N, Freitas KA, Chen YT, Shi Q, Valencia AM, Shifrut E, Kale N, Yost KE, Duffy CV, Daniel B, Hwee MA, Miao Z, Ashworth A, Mackall CL, Marson A, Carnevale J, Vardhana SA, Satpathy AT. Genome-wide CRISPR screens of T cell exhaustion identify chromatin remodeling factors that limit T cell persistence. Cancer Cell. 2022;40:768–786. doi: 10.1016/j.ccell.2022.06.001. - DOI - PMC - PubMed
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

