DDX18 coordinates nucleolus phase separation and nuclear organization to control the pluripotency of human embryonic stem cells
- PMID: 39738032
- PMCID: PMC11685540
- DOI: 10.1038/s41467-024-55054-8
DDX18 coordinates nucleolus phase separation and nuclear organization to control the pluripotency of human embryonic stem cells
Abstract
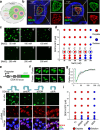
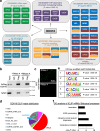
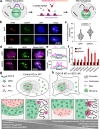
Pluripotent stem cells possess a unique nuclear architecture characterized by a larger nucleus and more open chromatin, which underpins their ability to self-renew and differentiate. Here, we show that the nucleolus-specific RNA helicase DDX18 is essential for maintaining the pluripotency of human embryonic stem cells. Using techniques such as Hi-C, DNA/RNA-FISH, and biomolecular condensate analysis, we demonstrate that DDX18 regulates nucleolus phase separation and nuclear organization by interacting with NPM1 in the granular nucleolar component, driven by specific nucleolar RNAs. Loss of DDX18 disrupts nucleolar substructures, impairing centromere clustering and perinucleolar heterochromatin (PNH) formation. To probe this further, we develop NoCasDrop, a tool enabling precise nucleolar targeting and controlled liquid condensation, which restores centromere clustering and PNH integrity while modulating developmental gene expression. This study reveals how nucleolar phase separation dynamics govern chromatin organization and cell fate, offering fresh insights into the molecular regulation of stem cell pluripotency.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Jerkovic, I. & Cavalli, G. Understanding 3D genome organization by multidisciplinary methods. Nat. Rev. Mol. Cell Biol. 22, 511–528 (2021). - PubMed
-
- Hug, C. B. & Vaquerizas, J. M. The birth of the 3D genome during early embryonic development. Trends Genet.34, 903–914 (2018). - PubMed
-
- Zheng, H. & Xie, W. The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol.20, 535–550 (2019). - PubMed
-
- Watanabe-Susaki, K. et al. Biosynthesis of ribosomal RNA in nucleoli regulates pluripotency and differentiation ability of pluripotent stem cells. Stem Cells32, 3099–3111 (2014). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- GM129157/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R21 HD106263/HD/NICHD NIH HHS/United States
- R01 HD095938/HD/NICHD NIH HHS/United States
- R35 GM154906/GM/NIGMS NIH HHS/United States
- R01 HD114122/HD/NICHD NIH HHS/United States
- R21 HD116446/HD/NICHD NIH HHS/United States
- R21HD116446/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- HD095938/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- R01 HD097268/HD/NICHD NIH HHS/United States
- HD097268/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- HD114122/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- R01 GM129157/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

