Genome-wide identification of TAL1's functional targets: insights into its mechanisms of action in primary erythroid cells
- PMID: 20566737
- PMCID: PMC2909570
- DOI: 10.1101/gr.104935.110
Genome-wide identification of TAL1's functional targets: insights into its mechanisms of action in primary erythroid cells
Abstract
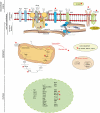
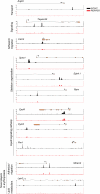
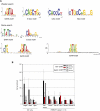
Coordination of cellular processes through the establishment of tissue-specific gene expression programs is essential for lineage maturation. The basic helix-loop-helix hemopoietic transcriptional regulator TAL1 (formerly SCL) is required for terminal differentiation of red blood cells. To gain insight into TAL1 function and mechanisms of action in erythropoiesis, we performed ChIP-sequencing and gene expression analyses from primary fetal liver erythroid cells. We show that TAL1 coordinates expression of genes in most known red cell-specific processes. The majority of TAL1's genomic targets require direct DNA-binding activity. However, one-fifth of TAL1's target sequences, mainly among those showing high affinity for TAL1, can recruit the factor independently of its DNA binding activity. An unbiased DNA motif search of sequences bound by TAL1 identified CAGNTG as TAL1-preferred E-box motif in erythroid cells. Novel motifs were also characterized that may help distinguish activated from repressed genes and suggest a new mechanism by which TAL1 may be recruited to DNA. Finally, analysis of recruitment of GATA1, a protein partner of TAL1, to sequences occupied by TAL1 suggests that TAL1's binding is necessary prior or simultaneous to that of GATA1. This work provides the framework to study regulatory networks leading to erythroid terminal maturation and to model mechanisms of action of tissue-specific transcription factors.
Figures



References
-
- Ader I, Brizuela L, Bouquerel P, Malavaud B, Cuvillier O 2008. Sphingosine kinase 1: A new modulator of hypoxia inducible factor 1α during hypoxia in human cancer cells. Cancer Res 68: 8635–8642 - PubMed
-
- Anderson KP, Crable SC, Lingrel JB 1998. Multiple proteins binding to a GATA-E box-GATA motif regulate the erythroid Kruppel-like factor (EKLF) gene. J Biol Chem 273: 14347–14354 - PubMed
-
- Anderson KP, Crable SC, Lingrel JB 2000. The GATA-E box-GATA motif in the EKLF promoter is required for in vivo expression. Blood 95: 1652–1655 - PubMed
-
- Aoki J, Nagai Y, Hosono H, Inoue K, Arai H 2002. Structure and function of phosphatidylserine-specific phospholipase A1. Biochim Biophys Acta 1582: 26–32 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
