Epigenetic memory of radiotherapy in dermal fibroblasts impairs wound repair capacity in cancer survivors
- PMID: 39468077
- PMCID: PMC11519383
- DOI: 10.1038/s41467-024-53295-1
Epigenetic memory of radiotherapy in dermal fibroblasts impairs wound repair capacity in cancer survivors
Abstract
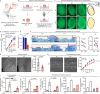
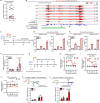
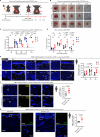
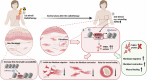
Radiotherapy (RT), a common cancer treatment, unintentionally harms surrounding tissues, including the skin, and hinders wound healing years after treatment. This study aims to understand the mechanisms behind these late-onset adverse effects. We compare skin biopsies from previously irradiated (RT+) and non-irradiated (RT-) sites in breast cancer survivors who underwent RT years ago. Here we show that the RT+ skin has compromised healing capacity and fibroblast functions. Using ATAC-seq, we discover altered chromatin landscapes in RT+ fibroblasts, with THBS1 identified as a crucial epigenetically primed wound repair-related gene. This is further confirmed by single-cell RNA-sequencing and spatial transcriptomic analysis of human wounds. Notably, fibroblasts in both murine and human post-radiation wound models show heightened and sustained THBS1 expression, impairing fibroblast motility and contractility. Treatment with anti-THBS1 antibodies promotes ex vivo wound closure in RT+ skin from breast cancer survivors. Our findings suggest that fibroblasts retain a long-term radiation memory in the form of epigenetic changes. Targeting this maladaptive epigenetic memory could mitigate RT's late-onset adverse effects, improving the quality of life for cancer survivors.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Thariat, J., Hannoun-Levi, J. M., Sun Myint, A., Vuong, T. & Gerard, J. P. Past, present, and future of radiotherapy for the benefit of patients. Nat. Rev. Clin. Oncol.10, 52–60 (2013). - PubMed
-
- Stone, H. B., Coleman, C. N., Anscher, M. S. & McBride, W. H. Effects of radiation on normal tissue: consequences and mechanisms. Lancet Oncol.4, 529–536 (2003). - PubMed
-
- Gieringer, M., Gosepath, J. & Naim, R. Radiotherapy and wound healing: principles, management and prospects (review). Oncol. Rep.26, 299–307 (2011). - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- Grant No. SSM2014-4016/Strålsäkerhetsmyndigheten (Radiation Safety Authority)
- No. 384224/151708/Gouvernement du Canada | Instituts de Recherche en Santé du Canada | Institute of Musculoskeletal Health and Arthritis (Institut de l'Appareil Locomoteur et de l'Arthrite)
- No. 161072/Radiumhemmets Forskningsfonder (Cancer Research Foundations of Radiumhemmet)
- No. SLS-886621/Svenska Läkaresällskapet (Swedish Society of Medicine)
- No. FoUI-962332/Stockholms Läns Landsting (Stockholm County Council)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

