Nutrigenomic regulation of sensory plasticity
- PMID: 36951889
- PMCID: PMC10036121
- DOI: 10.7554/eLife.83979
Nutrigenomic regulation of sensory plasticity
Abstract
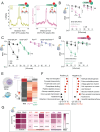
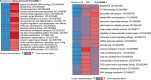
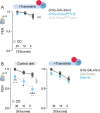
Diet profoundly influences brain physiology, but how metabolic information is transmuted into neural activity and behavior changes remains elusive. Here, we show that the metabolic enzyme O-GlcNAc Transferase (OGT) moonlights on the chromatin of the D. melanogaster gustatory neurons to instruct changes in chromatin accessibility and transcription that underlie sensory adaptations to a high-sugar diet. OGT works synergistically with the Mitogen Activated Kinase/Extracellular signal Regulated Kinase (MAPK/ERK) rolled and its effector stripe (also known as EGR2 or Krox20) to integrate activity information. OGT also cooperates with the epigenetic silencer Polycomb Repressive Complex 2.1 (PRC2.1) to decrease chromatin accessibility and repress transcription in the high-sugar diet. This integration of nutritional and activity information changes the taste neurons' responses to sugar and the flies' ability to sense sweetness. Our findings reveal how nutrigenomic signaling generates neural activity and behavior in response to dietary changes in the sensory neurons.
Keywords: D. melanogaster; gene regulation; genetics; genomics; neuroscience; nutrition; sensory neuroscience.
© 2023, Sung, Vaziri et al.
Conflict of interest statement
HS, AV, DW, RW, LF, MD No competing interests declared
Figures









Update of
- doi: 10.1101/2021.12.17.473205
References
-
- Andrews, Simon, and Others . FastQC: A Quality Control Tool for High Throughput Sequence Data. Cambridge, United Kingdom: Babraham Bioinformatics, Babraham Institute; 2010.
-
- Ardiel EL, McDiarmid TA, Timbers TA, Lee KCY, Safaei J, Pelech SL, Rankin CH. CaMK (CMK-1) and O-GlcNAc Transferase (OGT-1) Modulate Mechanosensory Responding and Habituation in an Interstimulus Interval-Dependent Manner in Caenorhabditis elegans. bioRxiv. 2018 doi: 10.1101/115972. - DOI
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- K99 DK128539/DK/NIDDK NIH HHS/United States
- R01DK130875/DK/NIDDK NIH HHS/United States
- R00 DK-97141/DK/NIDDK NIH HHS/United States
- R35GM128637/GM/NIGMS NIH HHS/United States
- P30 DK089503/DK/NIDDK NIH HHS/United States
- R35 GM128637/GM/NIGMS NIH HHS/United States
- R00 DK097141/DK/NIDDK NIH HHS/United States
- DK128539/DK/NIDDK NIH HHS/United States
- DP2DK-113750/DK/NIDDK NIH HHS/United States
- R01 DK130875/DK/NIDDK NIH HHS/United States
- T32 DA007268/DA/NIDA NIH HHS/United States
- DP2 DK113750/DK/NIDDK NIH HHS/United States
- T32 DA007268/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

