Loss of BRD4 induces cell senescence in HSC/HPCs by deregulating histone H3 clipping
- PMID: 37650863
- PMCID: PMC10561362
- DOI: 10.15252/embr.202357032
Loss of BRD4 induces cell senescence in HSC/HPCs by deregulating histone H3 clipping
Abstract
Bromodomain-containing protein 4 (BRD4) is overexpressed and functionally implicated in various myeloid malignancies. However, the role of BRD4 in normal hematopoiesis remains largely unknown. Here, utilizing an inducible Brd4 knockout mouse model, we find that deletion of Brd4 (Brd4Δ/Δ ) in the hematopoietic system impairs hematopoietic stem cell (HSC) self-renewal and differentiation, which associates with cell cycle arrest and senescence. ATAC-seq analysis shows increased chromatin accessibility in Brd4Δ/Δ hematopoietic stem/progenitor cells (HSC/HPCs). Genome-wide mapping with cleavage under target and release using nuclease (CUT&RUN) assays demonstrate that increased global enrichment of H3K122ac and H3K4me3 in Brd4Δ/Δ HSC/HPCs is associated with the upregulation of senescence-specific genes. Interestingly, Brd4 deletion increases clipped H3 (cH3) which correlates with the upregulation of senescence-specific genes and results in a higher frequency of senescent HSC/HPCs. Re-expression of BRD4 reduces cH3 levels and rescues the senescence rate in Brd4Δ/Δ HSC/HPCs. This study unveils an important role of BRD4 in HSC/HPC function by preventing H3 clipping and suppressing senescence gene expression.
Keywords: Brd4; hematopoiesis; histone clipping; senescence.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

- A
Kaplan–Meier survival curve representing percentage survival of Brd4 ∆/∆ mice after poly (I: C) injection. Log‐rank (Mantel–Cox) test; WT, n = 12; Brd4 ∆/+, n = 12; Brd4 ∆/∆, n = 12, P < 0.0001.
- B–D
Peripheral blood (PB) counts of RBC (B) (WT vs. Brd4 ∆/∆, P = 0.0026), Hb (C) (WT vs. Brd4 ∆/∆, P = 0.0016), and PLT (D) (WT vs. Brd4 ∆/∆, P = 0.0127) in WT, Brd4 ∆/+, and Brd4 ∆/∆ mice (n = 6 per genotype).
- E
Representative bone marrow from WT and Brd4 ∆/∆ mice.
- F
Bone marrow cellularity of WT, Brd4 ∆/+, and Brd4 ∆/∆ mice (n = 6 per genotype) (WT vs. Brd4 ∆/∆, P = 0.01).
- G
Hematoxylin and eosin (H&E)‐stained sections of femurs from representative WT and Brd4 ∆/∆ mice (14 days after poly (I: C) injection). Scale bar, 1 mm (Left), 10 μm (Right).
- H
Representative of May–Giemsa‐stained BM cytospins prepared from WT and Brd4 ∆/∆ mice. Scale bar, 100 μm.
- I
Flow cytometric analysis of erythroid cells in BM from WT and Brd4 ∆/∆ mice 14 days after first poly (I: C) injection.
- J
Flow cytometric analysis of myeloid cells in BM from WT and Brd4 ∆/∆ mice 14 days after first poly (I:C) injection.
- K
Frequency of Gr1−/Mac1+ cells in BM from WT and Brd4 ∆/∆ mice (WT, n = 4; Brd4 ∆/+, n = 3; Brd4 ∆/∆, n = 6) (WT vs. Brd4 ∆/∆, P < 0.0001) is shown.
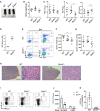
The gross appearance of representative mice from each genotype.
Body weight of WT, Brd4 ∆/+, and Brd4 ∆/∆ mice (14 days after poly (I: C) injection, n = 6) (WT vs. Brd4 ∆/∆, P = 0.28).
Peripheral blood (PB) counts of WBC (WT vs. Brd4 ∆/∆, P = 0.22), NE (WT vs. Brd4 ∆/∆, P = 0.1), and MO (WT vs Brd4 ∆/∆, P = 0.24) for WT, Brd4 ∆/+ and Brd4 ∆/∆ mice (n = 6 per genotype).
Quantitative analysis of M/E (myeloid cells to nucleated erythroid cells) ratio in WT and Brd4 ∆/∆ BM. Biological replicates, n = 3, unpaired Student's t‐test; ***P values < 0.001. Data are shown as the Mean ± S.E.M.
Flow cytometric analysis of T cells and B cells in BM from WT and Brd4 ∆/∆ mice 14 days after first poly (I: C) injection.
Spleen weight of WT, Brd4 ∆/+, and Brd4 ∆/∆ mice (14 days after poly (I: C) injection, n = 6) (WT vs. Brd4 ∆/∆, P = 0.015).
Spleen cellularity of WT, Brd4 ∆/+, and Brd4 ∆/∆ mice (14 days after poly (I: C) injection, n = 6) (WT vs. Brd4 ∆/∆, P = 0.42).
Hematoxylin and eosin (H&E)‐stained sections of spleens of representative WT and Brd4 ∆/∆ mice (14 days after poly (I: C) injection). Scale bar, 1 mm (Left), 10 μm (Right).
Flow cytometric analysis of LSK and LKS− populations in BM of representative WT, Brd4 ∆/+, and Brd4 ∆/∆ mice.
CFU‐C number per femur from WT and Brd4 ∆/∆ mice (n = 3). Data are presented as Mean ± S.E.M. ***P < 0.001.
HPP‐CFU assay using 3 × 104 bone marrow mononuclear cells from WT and Brd4 ∆/∆ mice (n = 6), ***P < 0.001.

- A
Flow cytometric analysis of Lin−, LT‐HSC (long‐term HSC), ST‐HSC (short‐term HSC), MPP (Multipotent progenitor), CMP (common myeloid progenitor), GMP (granulocyte/macrophage progenitor), and MEP (megakaryocyte‐erythrocyte progenitor) cell populations in BM of representative WT, Brd4 ∆/+, and Brd4 ∆/∆ mice.
- B, C
Quantitation of the percentage of HSC/HPC subpopulations in WT, Brd4 ∆/+, and Brd4 ∆/∆ mice (n = 6 per genotype). Lin−, WT vs. Brd4 ∆/∆, P < 0.0001; LKS−, WT vs. Brd4 ∆/∆, P < 0.0001; CMP, WT vs. Brd4 ∆/∆, P < 0.0001; GMP, WT vs. Brd4 ∆/∆, P = 0.0115; MEP, WT vs. Brd4 ∆/∆, P = 0.0102; LSK, WT vs. Brd4 ∆/∆, P = 0.0054; LT‐HSC, WT vs. Brd4 ∆/∆, P = 0.1153; ST‐HSC, WT vs. Brd4 ∆/∆, P = 0.0013; MPP, WT vs. Brd4 ∆/∆, P = 0.2682. Comparisons among the groups were formed by one‐way ANOVA.
- D
CFU‐C assay using 12,500 BMMNC cells from WT and Brd4 ∆/∆ mice. Biological replicates, n = 3, Group 50–500: P = 0.0619, Group > 500: P < 0.0001.
- E
CFU‐C assay using 100 LT‐HSC cells from WT and Brd4 ∆/∆ mice, n = 6, biological replicates, P < 0.001.
- F
Serial cell replating assays using BMMNC cells (3 mice per genotype, biological replicates). A total of 12,500 bone marrow mononuclear cells were used for each methylcellulose culture. The cells were replated weekly for 3 weeks.
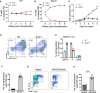
- A, B
The percentages of donor‐derived WT (n = 5, biological replicates) (A) or Brd4 ∆/∆ (n = 8, biological replicates) (B) CD45.2+ cells versus CD45.1+ cells in the peripheral blood of recipient animals at indicated time points are shown.
- C
Cell number counting for the liquid culture of LK cells from WT and Brd4 ∆/∆ mice. Biological replicates, n = 3, P < 0.0001.
- D
Flow cytometric analysis of myeloid cells in LK from WT and Brd4 ∆/∆ mice 7 days after liquid culture.
- E
Quantitation of the cell cycle in WT and Brd4 ∆/∆ mice. Biological duplicates, n = 3, G0/G1, P = 0.0001; S, P = 0.00017.
- F
Quantitation of the number of senescence cells per 100 LK cells from WT and Brd4 ∆/∆ mice. Biological replicates, n = 5, P < 0.0001.
- G
Flow cytometric analysis of cell cycle for 32D cells with or without ARV825 treatment.
- H
Quantitation of the number of senescence cells per 100 32D cells after 72 h of ARV825 (5 nM) treatment, biological replicates, n = 5, P < 0.0001.

Flow cytometric analysis of cell cycle for BM gated on Lin− from WT and Brd4 ∆/∆ mice.
Flow cytometric analysis of apoptotic cells in BM gated on Lin− from WT and Brd4 ∆/∆ mice 14 days after first poly (I: C) injection.
Senescence assay for LK cells. Representative of senescence staining for gated LK cells from BM. Scale bar, 10 μm.
Flow cytometry histograms showing SA‐beta Gal fluorescence in WT and Brd4 ∆/∆ HSC/HPCs, left to right: total BM, Lin− populations, LSK populations. Gray: Non‐staining, Blue: WT, Red: Brd4 ∆/∆.
The markers of senescence expression level in WT and Brd4 ∆/∆ HSC/HPCs. The mRNA level was detected by qPCR and normalized to GAPDH. Biological replicates, n = 3, unpaired Student's t‐test; ***P values < 0.001. Data are shown as the Mean ± S.E.M.
Representative of senescence staining for 32D cells after ARV825 treatment. Scale bar, 10 μm.
Flow cytometric analysis of apoptosis for 32D cells with or without 3 days ARV825 treatment.
Flow cytometry histograms showing SA‐beta Gal fluorescence in 32D cells with or without ARV‐825 treatment after 7 days culture, Gray: non‐staining, Blue: DMSO, Red: ARV825 treatment.
The markers of senescence expression level in 32D cells with or without ARV825 treatment after 7 days of culture. The mRNA level was detected by qPCR and normalized to GAPDH. Biological replicates, n = 3, unpaired Student's t‐test; ***P values < 0.001. Data are shown as the Mean ± S.E.M.
Flow cytometric analysis of apoptosis for 32D cells with or without ARV825 treatment after 7 days culture.

Heatmap of RNA‐seq analysis shows the up‐ and downregulated genes in Brd4 ∆/∆ versus WT LK cells (FDR < 0.05 and fold change in log > 1).
Gene set enrichment analysis (GSEA) shows that genes involved in the regulation of senescence are upregulated in Brd4 ∆/∆ LK cells. The normalized enrichment score (NES) and FDR are shown.
Uniform manifold approximation and projection (UMAP) visualization of HSC/HPC clusters identified from WT and Brd4 ∆/∆ cKit+ cells. Each dot represents one cell, and cluster identity is color coded (Seurat). Left: Overlap of WT and Brd4 ∆/∆. Right: the cluster representative of the UMAP visualization.
Violin plot of stemness transcription signature in different subpopulations, Mann–Whitney U‐test.
Violin plot of myeloid transcription signature in different subpopulations, Mann–Whitney U‐test.
GSEA for gene sets of senescence in HSPC population in Brd4 ∆/∆ cells versus WT cells. The colors reflect scaled NES, representing the degree of expression change. The size of the circle represents the FDR value.
UMAP visualization of HSC/HPC clusters identified from WT and Brd4 ∆/∆ cKit+ cells, colored by the signature score values of senescence gene sets.
GSEA for gene sets of HSC and cell cycle in the HSPC population in Brd4 ∆/∆ cells versus WT cells. The colors reflect scaled NES, representing the degree of expression change. The size of the circle depicts the FDR value.

- A
Gene set enrichment analysis (GSEA) shows that genes involved in the regulation of senescence are upregulated in Brd4 ∆/∆ LK cells. The normalized enrichment score (NES) and FDR are shown.
- B, C
Gene set enrichment analysis (GSEA) shows that genes are involved in the regulation of senescence from Ref. Saul et al (2022) (B) and Ref. Midha et al (2021) (C) are upregulated in Brd4 ∆/∆ LK cells. The normalized enrichment score (NES) and FDR are shown.
- D
Venn diagram depicts the number of differential cH3 peaks identified between control and Brd4 ∆/∆ LK cells associated with H3K27ac‐enriched differential peaks identified between control and Brd4 ∆/∆ LK cells.
- E
Venn diagram depicts the number of differential cH3 peaks identified between control and Brd4 ∆/∆ LK cells associated with H3K27me3‐enriched differential peaks identified between control and Brd4 ∆/∆ LK cells.
- F
Violin plot of mean scores within TSS ± 5 kb of cH3 CUT&RUN signals in senescence genes from diverse models in WT and Brd4 ∆/∆ LK cells. Left to right: Reactome senescence‐associated secretory phenotype SASP, gene list from Ref. Saul et al (2022), gene list from Ref. Midha et al (2021).

Heatmap of genome‐wide ATAC‐seq signal profile around TSS (± 2,000 bp).
Heatmap of accessibility signal profile among increased and decreased peak regions.
Functional enrichment analysis of genes with increased ATAC‐seq peaks in Brd4 ∆/∆ compared with WT LK cells.
Functional enrichment analysis of genes with decreased ATAC‐seq peaks in Brd4 ∆/∆ compared with WT LK cells.
Footprinting analysis with TOBIAS illustrates global changes in transcription factor footprint depth in Brd4 ∆/∆ versus WT LK cells. Each dot represents one transcription factor (TF). WT higher TFs are labeled as blue, and Brd4 ∆/∆ higher TFs are labeled as red.
The intersection of RNA‐seq and ATAC‐seq in LK cells. Promoter peak log2FoldChange scores from ATAC‐seq analysis (FDR < 0.05) and log2FoldChange scores from RNA‐seq analysis (DEGs) were tabulated and gene matched. Pearson's product–moment correlation test, P‐value < 2.2e‐16, R coefficient score = 0.45.
Heatmap displays the densities of ATAC‐seq peaks on 42 senescence‐activated genes (TSS ± 3 kb). P‐value = 3.969e‐08.
ATAC‐seq tracks show chromatin accessibility at the S100a8 and S100a9 genes with RNA‐seq reads coverage in WT and Brd4 ∆/∆ LK cells. Regions showing increased accessibility and increased gene expression in Brd4 ∆/∆ LK cells.
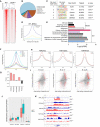
BRD4 CUT&RUN signal in WT and Brd4 ∆/∆ HSC/HPCs.
Pie chart showing the distribution of BRD4‐binding sites across genomic regions in HSC/HPCs.
De novo DNA sequence motifs identified in BRD4‐bound regions.
Meta‐gene tracks of BRD4 CUT&RUN signal averaged over all promoter‐TSS tracks grouped by relative expression levels (highly expressed, TPM ≥ 1; lowly expressed, TPM < 1). X‐axis, base pairs relative to TSS; Y‐axis, BRD4 CUT&RUN signal intensity.
Enrichment analysis of genes with BRD4 bound in WT cells from KEGG and gene ontology database.
The signal correlation of BRD4 with other histone modifications from WT LK cells.
Correlation score of BRD4 with other histone modifications.
Global levels of histone modifications at peaks and flanking 3‐kb regions. For each comparison, normalized coverages by sequencing depth were scaled to 100% and averaged in two biological replicates.
Scatter plots with linear fit show correlations between changes in H3K27ac, H3K122ac, and H3K4me3, and changes in gene expression in WT and Brd4 ∆/∆ LK cells. Pearson's correlation coefficient R‐values are shown, P‐value < 2.2e‐16 for all correlation tests. X‐ and Y‐axes show the log2‐transformed fold change (log2FC) for each histone enrichment and mRNA expression level, respectively. DEGs are marked in red.
Boxplot of mean scores of histone modification CUT&RUN signals in 42 senescence‐activated genes in WT and Brd4 ∆/∆ LK cells (TSS ± 5 kb). The bottom of the lower whisker, the bottom of the box, the middle band, the top of the box, and the top of the upper whisker represent the minimum, first quantile, median, third quantile, and maximum of histone modification levels on the 42 genes, respectively. The average of two biological replicates was used to quantify the level of specific histone modification level on each of the 42 senescence‐associated genes.
Representative genome browser tracks showing H3K27ac, H3K122ac, H3K4me3, and H3K27me3 enrichment on the S100a8 gene.

Western blot analysis of WT and Brd4 ∆/∆ HSC/HPC nuclear protein with the indicated histone PTM‐specific antibodies. Arrows indicate the cleavage product. The BRD4 level is also shown in Appendix Fig S1 using the same image.
H3 cleavage assay of WT and Brd4 ∆/∆ HSPC cell lysate was analyzed by western blot with BRD4 and Streptavidin antibodies. Arrows indicated the cleavage H3.3.
H3 cleavage assay of WT and Brd4 ∆/∆ HSPC cell lysate with or without ELANE inhibitor (ELANEi) were analyzed by western blot with BRD4 and Streptavidin antibodies. Arrows indicated the cleavage H3.3.
WT and Brd4 ∆/∆ HSC/HPC cells were fractionated into chromatin‐free (cytoplasmic and nuclear soluble) and chromatin fractions and blotted by indicated antibodies. Arrows indicate the cleavage H3.
Association between cH3 enrichment and active transcription. The cH3 levels at the TSS‐proximal regions of the genes grouped with relative expression level (highly expressed, TPM ≥ 1; lowly expressed, TPM < 1). X‐axis, base pairs relative to TSS; Y‐axis, cH3 CUT&RUN signal intensity.
Enrichment analysis of genes with cH3 occupancy in Brd4 ∆/∆ HSC/HPC cells from KEGG and GO database.
Venn diagram depicts the number of cH3 peaks‐related genes identified in Brd4 ∆/∆ LK cells associated with accessible (ATAC‐seq) peaks‐related genes identified in Brd4 ∆/∆ LK cells.
Venn diagram depicts the number of differential cH3 peaks‐related genes identified between control and Brd4 ∆/∆ LK cells associated with BRD4‐enriched peaks‐related genes identified in control LK cells.
Venn diagram depicts the number of differential cH3 peaks‐related genes identified between control and Brd4 ∆/∆ LK cells associated with H3K122ac‐enriched differential peaks‐related genes identified between control and Brd4 ∆/∆ LK cells.
Venn diagrams show the overlap among differential cH3_BRD4, cH3_H3K122ac, and cH3_H3K27ac peaks‐related genes.
Violin plot of mean scores within TSS ± 5 kb of cH3 CUT&RUN signals in 42 senescence‐activated genes in WT and Brd4 ∆/∆ LK cells. Unpaired Student's t‐test, P‐value = 8.866e−10.
ChIP‐quantitative PCR for cH3 at senescence genes in BRD4 WT and Brd4 ∆/∆ HSC/HPC cells. Biological replicates: n = 3, Mean ± S.E.M; one‐way ANOVA tests; P‐values < 0.001 (cH3‐Brd4 ∆/∆ vs. cH3‐WT).

Quantification of β‐gal+ cells per 500 cells in 32D cells expressing the indicated histones; Mean ± S.E.M. (n = 5 biological replicates); Unpaired Student's t‐test; ***P‐values < 0.001 (cH3.3 vs. H3.3).
Quantification of colony numbers per 100 GFP+ cells for colony formation assay with 32D cells expressing the indicated histones. Colony size > 500 cells, ***P‐values = 0.0005; colony size from 50 to 500 cells, P‐values = 0.35. Mean ± S.E.M. (n = 5 biological replicates); unpaired Student's t‐test.
Western blot analysis of nuclear protein from WT, Brd4 ∆/∆, and Brd4 OE ; Brd4 ∆/∆ HSC/HPCs with the indicated antibodies. Arrows indicated the cH3 product.
Representative of senescence staining for gated LK cells from Brd4 ∆/∆ and Brd4 OE ; Brd4 ∆/∆ mice, scale bar, 10 μm.
Quantitation of the frequency of senescence cells per 100 LK cells from Brd4 ∆/∆ and Brd4 OE ; Brd4 ∆/∆ mice. Biological replicates, n = 5, unpaired Student's t‐test; ***P‐values < 0.001. Data are shown as the Mean ± S.E.M.
CFU‐C assay using 2.5 × 104 BMMNC cells from WT, Brd4 ∆/∆ and Brd4 OE ; Brd4 ∆/∆ mice. Biological replicates, n = 3 unpaired Student's t‐test; ***P‐values < 0.001. Data are shown as the Mean ± S.E.M.
Comment in
-
Hematopoiesis: a BETter understanding.EMBO Rep. 2023 Oct 9;24(10):e57927. doi: 10.15252/embr.202357927. Epub 2023 Aug 31. EMBO Rep. 2023. PMID: 37650879 Free PMC article.
References
-
- Amorim S, Stathis A, Gleeson M, Iyengar S, Magarotto V, Leleu X, Morschhauser F, Karlin L, Broussais F, Rezai K et al (2016) Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose‐escalation, open‐label, pharmacokinetic, phase 1 study. Lancet Haematol 3: e196–e204 - PubMed
-
- Bansal H, Kornblau S, Qiu YH, Coombes KR, Panneerdoss P, Karnad A, Weitman S, Tomlinson G, Bansal S, Iyer SP (2017) Overexpression of BRD4 Is an adverse prognostic factor in acute myeloid leukemia. ASH Abstract 130: 3794
-
- Berthon C, Raffoux E, Thomas X, Vey N, Gomez‐Roca C, Yee K, Taussig DC, Rezai K, Roumier C, Herait P et al (2016) Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose‐escalation, phase 1 study. Lancet Haematol 3: e186–e195 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

