Cross-species hybridization of microarrays for studying tumor transcriptome of brain metastasis
- PMID: 21987811
- PMCID: PMC3198333
- DOI: 10.1073/pnas.1114210108
Cross-species hybridization of microarrays for studying tumor transcriptome of brain metastasis
Abstract
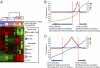
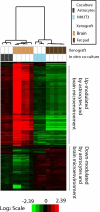
Although the importance of the cellular microenvironment (soil) during invasion and metastasis of cancer cells (seed) has been well-recognized, technical challenges have limited the ability to assess the influence of the microenvironment on cancer cells at the molecular level. Here, we show that an experimental strategy, competitive cross-species hybridization of microarray experiments, can characterize the influence of different microenvironments on cancer cells by independently extracting gene expression data of cancer and host cells when human cancer cells were xenografted into different organ sites of immunocompromised mice. Surprisingly, the analysis of gene expression data showed that the brain microenvironment induces complete reprogramming of metastasized cancer cells, resulting in a gain of neuronal cell characteristics and mimicking neurogenesis during development. We also show that epigenetic changes coincide with transcriptional reprogramming in cancer cells. These observations provide proof of principle for competitive cross-species hybridization of microarray experiments to characterize the effect of the microenvironment on tumor cell behavior.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Nguyen DX, Bos PD, Massagué J. Metastasis: From dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–284. - PubMed
-
- Freilich RJ, Seidman AD, DeAngelis LM. Central nervous system progression of metastatic breast cancer in patients treated with paclitaxel. Cancer. 1995;76:232–236. - PubMed
-
- Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. - PubMed
-
- Friedl P, Wolf K. Tumour-cell invasion and migration: Diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. - PubMed
-
- Finak G, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

