3' Untranslated regions mediate transcriptional interference between convergent genes both locally and ectopically in Saccharomyces cerevisiae
- PMID: 24465217
- PMCID: PMC3900390
- DOI: 10.1371/journal.pgen.1004021
3' Untranslated regions mediate transcriptional interference between convergent genes both locally and ectopically in Saccharomyces cerevisiae
Abstract
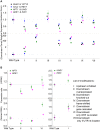
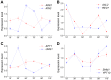
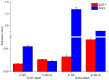
Paired sense and antisense (S/AS) genes located in cis represent a structural feature common to the genomes of both prokaryotes and eukaryotes, and produce partially complementary transcripts. We used published genome and transcriptome sequence data and found that over 20% of genes (645 pairs) in the budding yeast Saccharomyces cerevisiae genome are arranged in convergent pairs with overlapping 3'-UTRs. Using published microarray transcriptome data from the standard laboratory strain of S. cerevisiae, our analysis revealed that expression levels of convergent pairs are significantly negatively correlated across a broad range of environments. This implies an important role for convergent genes in the regulation of gene expression, which may compensate for the absence of RNA-dependent mechanisms such as micro RNAs in budding yeast. We selected four representative convergent gene pairs and used expression assays in wild type yeast and its genetically modified strains to explore the underlying patterns of gene expression. Results showed that convergent genes are reciprocally regulated in yeast populations and in single cells, whereby an increase in expression of one gene produces a decrease in the expression of the other, and vice-versa. Time course analysis of the cell cycle illustrated the functional significance of this relationship for the three pairs with relevant functional roles. Furthermore, a series of genetic modifications revealed that the 3'-UTR sequence plays an essential causal role in mediating transcriptional interference, which requires neither the sequence of the open reading frame nor the translation of fully functional proteins. More importantly, transcriptional interference persisted even when one of the convergent genes was expressed ectopically (in trans) and therefore does not depend on the cis arrangement of convergent genes; we conclude that the mechanism of transcriptional interference cannot be explained by the transcriptional collision model, which postulates a clash between simultaneous transcriptional processes occurring on opposite DNA strands.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Comment in
-
Transcription: Interference from near and far.Nat Rev Genet. 2014 Mar;15(3):144. doi: 10.1038/nrg3675. Epub 2014 Feb 11. Nat Rev Genet. 2014. PMID: 24514438 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

