Integrating genetic regulation and single-cell expression with GWAS prioritizes causal genes and cell types for glaucoma
- PMID: 38195602
- PMCID: PMC10776627
- DOI: 10.1038/s41467-023-44380-y
Integrating genetic regulation and single-cell expression with GWAS prioritizes causal genes and cell types for glaucoma
Abstract
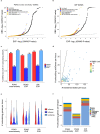
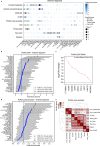
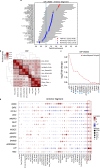
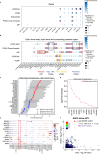
Primary open-angle glaucoma (POAG), characterized by retinal ganglion cell death, is a leading cause of irreversible blindness worldwide. However, its molecular and cellular causes are not well understood. Elevated intraocular pressure (IOP) is a major risk factor, but many patients have normal IOP. Colocalization and Mendelian randomization analysis of >240 POAG and IOP genome-wide association study (GWAS) loci and overlapping expression and splicing quantitative trait loci (e/sQTLs) in 49 GTEx tissues and retina prioritizes causal genes for 60% of loci. These genes are enriched in pathways implicated in extracellular matrix organization, cell adhesion, and vascular development. Analysis of single-nucleus RNA-seq of glaucoma-relevant eye tissues reveals that the POAG and IOP colocalizing genes and genome-wide associations are enriched in specific cell types in the aqueous outflow pathways, retina, optic nerve head, peripapillary sclera, and choroid. This study nominates IOP-dependent and independent regulatory mechanisms, genes, and cell types that may contribute to POAG pathogenesis.
© 2024. The Author(s).
Conflict of interest statement
J.R.S. was a consultant for Biogen, but none of this work benefits or benefited from that relationship. T.V.Z. is an employee of Regeneron. A.P.K. has acted as a paid consultant or lecturer to Abbvie, Aerie, Allergan, Google Health, Heidelberg Engineering, Novartis, Reichert, Santen and Thea. L.R.P. is a paid consultant to Twenty Twenty. St.M. is a co-founder of and holds stock in Seonix Pty Ltd. J.L.W. is a recent consultant for CRISPR Therapeutics and Editas. The remaining authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions

