PCLAF-DREAM drives alveolar cell plasticity for lung regeneration
- PMID: 39448571
- PMCID: PMC11502753
- DOI: 10.1038/s41467-024-53330-1
PCLAF-DREAM drives alveolar cell plasticity for lung regeneration
Abstract
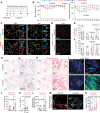
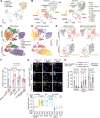
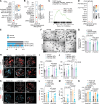
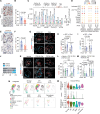
Cell plasticity, changes in cell fate, is crucial for tissue regeneration. In the lung, failure of regeneration leads to diseases, including fibrosis. However, the mechanisms governing alveolar cell plasticity during lung repair remain elusive. We previously showed that PCLAF remodels the DREAM complex, shifting the balance from cell quiescence towards cell proliferation. Here, we find that PCLAF expression is specific to proliferating lung progenitor cells, along with the DREAM target genes transactivated by lung injury. Genetic ablation of Pclaf impairs AT1 cell repopulation from AT2 cells, leading to lung fibrosis. Mechanistically, the PCLAF-DREAM complex transactivates CLIC4, triggering TGF-β signaling activation, which promotes AT1 cell generation from AT2 cells. Furthermore, phenelzine that mimics the PCLAF-DREAM transcriptional signature increases AT2 cell plasticity, preventing lung fibrosis in organoids and mice. Our study reveals the unexpected role of the PCLAF-DREAM axis in promoting alveolar cell plasticity, beyond cell proliferation control, proposing a potential therapeutic avenue for lung fibrosis prevention.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Chonghaile, M. N., Higgins, B. D., Costello, J. & Laffey, J. G. Hypercapnic acidosis attenuates lung injury induced by established bacterial pneumonia. Anesthesiology109, 837–848 (2008). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
- R01 HL130129/HL/NHLBI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- R03 CA256207/CA/NCI NIH HHS/United States
- R01 CA278967/CA/NCI NIH HHS/United States
- R00 HL155845/HL/NHLBI NIH HHS/United States
- R01 HL153511/HL/NHLBI NIH HHS/United States
- RP200315, RP180672, RP200504/Cancer Prevention and Research Institute of Texas (Cancer Prevention Research Institute of Texas)
- R03 CA279867/CA/NCI NIH HHS/United States
- CA193297, CA256207, CA278967, CA278971, CA279867, P50 CA83639, P30 CA016672, CA125123, RR024574/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R35 HL171346/HL/NHLBI NIH HHS/United States
- R01 CA193297/CA/NCI NIH HHS/United States
- P30 CA125123/CA/NCI NIH HHS/United States
- K99CA286761/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- S10 RR024574/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

