CTLA-4 expressing innate lymphoid cells modulate mucosal homeostasis in a microbiota dependent manner
- PMID: 39496592
- PMCID: PMC11535242
- DOI: 10.1038/s41467-024-51719-6
CTLA-4 expressing innate lymphoid cells modulate mucosal homeostasis in a microbiota dependent manner
Abstract
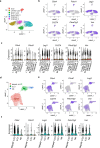
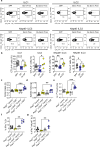
The maintenance of intestinal homeostasis is a fundamental process critical for organismal integrity. Sitting at the interface of the gut microbiome and mucosal immunity, adaptive and innate lymphoid populations regulate the balance between commensal micro-organisms and pathogens. Checkpoint inhibitors, particularly those targeting the CTLA-4 pathway, disrupt this fine balance and can lead to inflammatory bowel disease and immune checkpoint colitis. Here, we show that CTLA-4 is expressed by innate lymphoid cells and that its expression is regulated by ILC subset-specific cytokine cues in a microbiota-dependent manner. Genetic deletion or antibody blockade of CTLA-4 in multiple in vivo models of colitis demonstrates that this pathway plays a key role in intestinal homeostasis. Lastly, we have found that this observation is conserved in human IBD. We propose that this population of CTLA-4-positive ILC may serve as an important target for the treatment of idiopathic and iatrogenic intestinal inflammation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Guntermann, C. & Alexander, D. R. CTLA-4 suppresses proximal TCR signaling in resting human CD4+ T cells by inhibiting ZAP-70 Tyr319 phosphorylation: a potential role for tyrosine phosphatases. J. Immunol.168, 4420–4429 (2002). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

