RBFOX2 deregulation promotes pancreatic cancer progression and metastasis through alternative splicing
- PMID: 38114498
- PMCID: PMC10730836
- DOI: 10.1038/s41467-023-44126-w
RBFOX2 deregulation promotes pancreatic cancer progression and metastasis through alternative splicing
Abstract
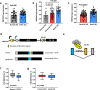
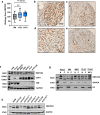
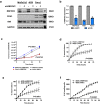
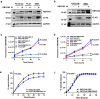
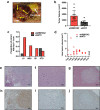
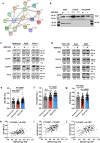
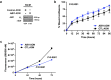
RNA splicing is an important biological process associated with cancer initiation and progression. However, the contribution of alternative splicing to pancreatic cancer (PDAC) development is not well understood. Here, we identify an enrichment of RNA binding proteins (RBPs) involved in splicing regulation linked to PDAC progression from a forward genetic screen using Sleeping Beauty insertional mutagenesis in a mouse model of pancreatic cancer. We demonstrate downregulation of RBFOX2, an RBP of the FOX family, promotes pancreatic cancer progression and liver metastasis. Specifically, we show RBFOX2 regulates exon splicing events in transcripts encoding proteins involved in cytoskeletal remodeling programs. These exons are differentially spliced in PDAC patients, with enhanced exon skipping in the classical subtype for several RBFOX2 targets. RBFOX2 mediated splicing of ABI1, encoding the Abelson-interactor 1 adapter protein, controls the abundance and localization of ABI1 protein isoforms in pancreatic cancer cells and promotes the relocalization of ABI1 from the cytoplasm to the periphery of migrating cells. Using splice-switching antisense oligonucleotides (AONs) we demonstrate the ABI1 ∆Ex9 isoform enhances cell migration. Together, our data identify a role for RBFOX2 in promoting PDAC progression through alternative splicing regulation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- figshare/10.6084/m9.figshare.24212682.
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

