Genomic predictors for recurrence patterns of hepatocellular carcinoma: model derivation and validation
- PMID: 25536056
- PMCID: PMC4275163
- DOI: 10.1371/journal.pmed.1001770
Genomic predictors for recurrence patterns of hepatocellular carcinoma: model derivation and validation
Abstract
Background: Typically observed at 2 y after surgical resection, late recurrence is a major challenge in the management of hepatocellular carcinoma (HCC). We aimed to develop a genomic predictor that can identify patients at high risk for late recurrence and assess its clinical implications.
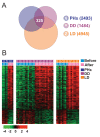
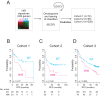
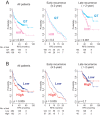
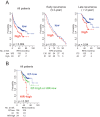
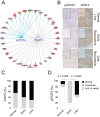
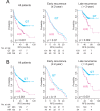
Methods and findings: Systematic analysis of gene expression data from human liver undergoing hepatic injury and regeneration revealed a 233-gene signature that was significantly associated with late recurrence of HCC. Using this signature, we developed a prognostic predictor that can identify patients at high risk of late recurrence, and tested and validated the robustness of the predictor in patients (n = 396) who underwent surgery between 1990 and 2011 at four centers (210 recurrences during a median of 3.7 y of follow-up). In multivariate analysis, this signature was the strongest risk factor for late recurrence (hazard ratio, 2.2; 95% confidence interval, 1.3-3.7; p = 0.002). In contrast, our previously developed tumor-derived 65-gene risk score was significantly associated with early recurrence (p = 0.005) but not with late recurrence (p = 0.7). In multivariate analysis, the 65-gene risk score was the strongest risk factor for very early recurrence (<1 y after surgical resection) (hazard ratio, 1.7; 95% confidence interval, 1.1-2.6; p = 0.01). The potential significance of STAT3 activation in late recurrence was predicted by gene network analysis and validated later. We also developed and validated 4- and 20-gene predictors from the full 233-gene predictor. The main limitation of the study is that most of the patients in our study were hepatitis B virus-positive. Further investigations are needed to test our prediction models in patients with different etiologies of HCC, such as hepatitis C virus.
Conclusions: Two independently developed predictors reflected well the differences between early and late recurrence of HCC at the molecular level and provided new biomarkers for risk stratification. Please see later in the article for the Editors' Summary.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- International Agency for Research on Cancer (2013) GLOBOCAN 2012: estimated cancer incidence, mortality and prevalence worldwide in 2012. Lyon (France): International Agency for Research on Cancer.
-
- Davila JA, Morgan RO, Shaib Y, McGlynn KA, El Serag HB (2004) Hepatitis C infection and the increasing incidence of hepatocellular carcinoma: a population-based study. Gastroenterology 127: 1372–1380. - PubMed
-
- El Serag HB, Mason AC (1999) Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med 340: 745–750. - PubMed
-
- El Serag HB (2004) Hepatocellular carcinoma: recent trends in the United States. Gastroenterology 127: S27–S34. - PubMed
-
- Bruix J, Llovet JM (2002) Prognostic prediction and treatment strategy in hepatocellular carcinoma. Hepatology 35: 519–524. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

