MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L
- PMID: 21741597
- PMCID: PMC3329803
- DOI: 10.1016/j.ccr.2011.06.010
MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L
Abstract
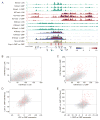
The histone 3 lysine 79 (H3K79) methyltransferase Dot1l has been implicated in the development of leukemias bearing translocations of the Mixed Lineage Leukemia (MLL) gene. We identified the MLL-fusion targets in an MLL-AF9 leukemia model, and conducted epigenetic profiling for H3K79me2, H3K4me3, H3K27me3, and H3K36me3 in hematopoietic progenitor and leukemia stem cells (LSCs). We found abnormal profiles only for H3K79me2 on MLL-AF9 fusion target loci in LSCs. Inactivation of Dot1l led to downregulation of direct MLL-AF9 targets and an MLL translocation-associated gene expression signature, whereas global gene expression remained largely unaffected. Suppression of MLL translocation-associated gene expression corresponded with dependence of MLL-AF9 leukemia on Dot1l in vivo. These data point to DOT1L as a potential therapeutic target in MLL-rearranged leukemia.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures





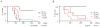
References
-
- Albert M, Helin K. Histone methyltransferases in cancer. Semin Cell Dev Biol. 2010;21:209–220. - PubMed
-
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. - PubMed
-
- Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. - PubMed
-
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

