Inhibition of GPX4 enhances CDK4/6 inhibitor and endocrine therapy activity in breast cancer
- PMID: 39500869
- PMCID: PMC11538343
- DOI: 10.1038/s41467-024-53837-7
Inhibition of GPX4 enhances CDK4/6 inhibitor and endocrine therapy activity in breast cancer
Abstract
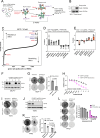
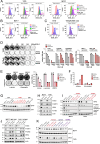
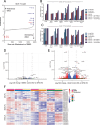
CDK4/6 inhibition in combination with endocrine therapy is the standard of care for estrogen receptor (ER+) breast cancer, and although cytostasis is frequently observed, new treatment strategies that enhance efficacy are required. Here, we perform two independent genome-wide CRISPR screens to identify genetic determinants of CDK4/6 and endocrine therapy sensitivity. Genes involved in oxidative stress and ferroptosis modulate sensitivity, with GPX4 as the top sensitiser in both screens. Depletion or inhibition of GPX4 increases sensitivity to palbociclib and giredestrant, and their combination, in ER+ breast cancer models, with GPX4 null xenografts being highly sensitive to palbociclib. GPX4 perturbation additionally sensitises triple negative breast cancer (TNBC) models to palbociclib. Palbociclib and giredestrant induced oxidative stress and disordered lipid metabolism, leading to a ferroptosis-sensitive state. Lipid peroxidation is promoted by a peroxisome AGPAT3-dependent pathway in ER+ breast cancer models, rather than the classical ACSL4 pathway. Our data demonstrate that CDK4/6 and ER inhibition creates vulnerability to ferroptosis induction, that could be exploited through combination with GPX4 inhibitors, to enhance sensitivity to the current therapies in breast cancer.
© 2024. The Author(s).
Conflict of interest statement
J.C., J-P.F., P.P., A.B., M.R.C, J.G., Q.L., W.R.W., W.S., J.V., M.H., and C.M. are or were employees of Genentech/Roche and hold Roche shares. All other Authors declare no competing interests.
Figures




References
-
- Goetz, M. P. et al. MONARCH 3: Abemaciclib as initial therapy for advanced breast cancer. J. Clin. Oncol.35, 3638–3646 (2017). - PubMed
-
- Hortobagyi, G. N. et al. Ribociclib as first-line therapy for HR-Positive, advanced breast cancer. N. Engl. J. Med.375, 1738–1748 (2016). - PubMed
-
- Finn, R. S. et al. Palbociclib and Letrozole in advanced breast cancer. N. Engl. J. Med.375, 1925–1936 (2016). - PubMed
-
- Tolaney, S. M. et al. Abemaciclib plus trastuzumab with or without fulvestrant versus trastuzumab plus standard-of-care chemotherapy in women with hormone receptor-positive, HER2-positive advanced breast cancer (monarcHER): a randomised, open-label, phase 2 trial. Lancet Oncol.21, 763–775 (2020). - PubMed
Publication types
MeSH terms
Substances
Associated data
- SRA/PRJNA989103
- Actions
LinkOut - more resources
Full Text Sources
Medical

