CDK9 recruits HUWE1 to degrade RARα and offers therapeutic opportunities for cutaneous T-cell lymphoma
- PMID: 39632829
- PMCID: PMC11618697
- DOI: 10.1038/s41467-024-54354-3
CDK9 recruits HUWE1 to degrade RARα and offers therapeutic opportunities for cutaneous T-cell lymphoma
Abstract
Cutaneous T-cell lymphoma (CTCL) is a heterogeneous non-Hodgkin lymphoma originating in the skin and invading the systemic hematopoietic system. Current treatments, including chemotherapy and monoclonal antibodies yielded limited responses with high incidence of side effects, highlighting the need for targeted therapy. Screening with small inhibitors library, herein we identify cyclin dependent kinase 9 (CDK9) as a driver of CTCL growth. Single-cell RNA-seq analysis reveals a CDK9high malignant T cell cluster with a unique actively proliferating feature. Inhibition, depletion or proteolysis targeting chimera (PROTAC)-mediated degradation of CDK9 significantly reduces CTCL cell growth in vitro and in murine models. CDK9 also promotes degradation of retinoic acid receptor α (RARα) via recruiting the E3 ligase HUWE1. Co-administration of CDK9-PROTAC (GT-02897) with all-trans retinoic acid (ATRA) leads to synergistic attenuation of tumor growth in vitro and in xenograft models, providing a potential translational treatment for complete eradication of CTCL.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







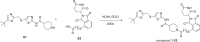

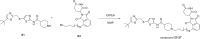
References
-
- Whittaker, S., Hoppe, R. & Prince, H. M. How I treat mycosis fungoides and Sezary syndrome. Blood127, 3142–3153 (2016). - PubMed
-
- Dummer, R. et al. Cutaneous T cell lymphoma. Nat. Rev. Dis. Prim.7, 61 (2021). - PubMed
-
- Kamijo, H. et al. Aberrant CD137 ligand expression induced by GATA6 overexpression promotes tumor progression in cutaneous T-cell lymphoma. Blood132, 1922–1935 (2018). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

