Breast cancer methylomes establish an epigenomic foundation for metastasis
- PMID: 21430268
- PMCID: PMC3146366
- DOI: 10.1126/scitranslmed.3001875
Breast cancer methylomes establish an epigenomic foundation for metastasis
Abstract
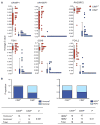
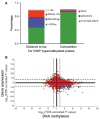
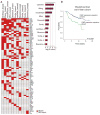
Cancer-specific alterations in DNA methylation are hallmarks of human malignancies; however, the nature of the breast cancer epigenome and its effects on metastatic behavior remain obscure. To address this issue, we used genome-wide analysis to characterize the methylomes of breast cancers with diverse metastatic behavior. Groups of breast tumors were characterized by the presence or absence of coordinate hypermethylation at a large number of genes, demonstrating a breast CpG island methylator phenotype (B-CIMP). The B-CIMP provided a distinct epigenomic profile and was a strong determinant of metastatic potential. Specifically, the presence of the B-CIMP in tumors was associated with low metastatic risk and survival, and the absence of the B-CIMP was associated with high metastatic risk and death. B-CIMP loci were highly enriched for genes that make up the metastasis transcriptome. Methylation at B-CIMP genes accounted for much of the transcriptomal diversity between breast cancers of varying prognosis, indicating a fundamental epigenomic contribution to metastasis. Comparison of the loci affected by the B-CIMP with those affected by the hypermethylator phenotype in glioma and colon cancer revealed that the CIMP signature was shared by multiple human malignancies. Our data provide a unifying epigenomic framework linking breast cancers with varying outcome and transcriptomic changes underlying metastasis. These findings significantly enhance our understanding of breast cancer oncogenesis and aid the development of new prognostic biomarkers for this common malignancy.
Conflict of interest statement
Figures




References
-
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K. Cancer Genome Atlas Research Network, Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. - PMC - PubMed
-
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. - PubMed
-
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. - PubMed
-
- Saal LH, Gruvberger-Saal SK, Persson C, Lövgren K, Jumppanen M, Staaf J, Jönsson G, Pires MM, Maurer M, Holm K, Koujak S, Subramaniyam S, Vallon-Christersson J, Olsson H, Su T, Memeo L, Ludwig T, Ethier SP, Krogh M, Szabolcs M, Murty VV, Isola J, Hibshoosh H, Parsons R, Borg A. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008;40:102–107. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

