AP-2γ regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription
- PMID: 21572391
- PMCID: PMC3155293
- DOI: 10.1038/emboj.2011.151
AP-2γ regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription
Erratum in
- EMBO J. 2011;30(13):2750. Sung, Win King [corrected to Sung, Wing Kin]
Abstract
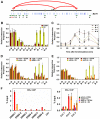
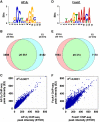
Oestrogen receptor α (ERα) is key player in the progression of breast cancer. Recently, the cistrome and interactome of ERα were mapped in breast cancer cells, revealing the importance of spatial organization in oestrogen-mediated transcription. However, the underlying mechanism of this process is unclear. Here, we show that ERα binding sites (ERBS) identified from the Chromatin Interaction Analysis-Paired End DiTag of ERα are enriched for AP-2 motifs. We demonstrate the transcription factor, AP-2γ, which has been implicated in breast cancer oncogenesis, binds to ERBS in a ligand-independent manner. Furthermore, perturbation of AP-2γ expression impaired ERα DNA binding, long-range chromatin interactions, and gene transcription. In genome-wide analyses, we show that a large number of AP-2γ and ERα binding events converge together across the genome. The majority of these shared regions are also occupied by the pioneer factor, FoxA1. Molecular studies indicate there is functional interplay between AP-2γ and FoxA1. Finally, we show that most ERBS associated with long-range chromatin interactions are colocalized with AP-2γ and FoxA1. Together, our results suggest AP-2γ is a novel collaborative factor in ERα-mediated transcription.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Ali S, Coombes RC (2000) Estrogen receptor alpha in human breast cancer: occurrence and significance. J Mammary Gland Biol Neoplasia 5: 271–281 - PubMed
-
- Arighi E, Borrello MG, Sariola H (2005) RET tyrosine kinase signaling in development and cancer. Cytokine Growth Factor Rev 16: 441–467 - PubMed
-
- Boulay A, Breuleux M, Stephan C, Fux C, Brisken C, Fiche M, Wartmann M, Stumm M, Lane HA, Hynes NE (2008) The Ret receptor tyrosine kinase pathway functionally interacts with the ERalpha pathway in breast cancer. Cancer Res 68: 3743–3751 - PubMed
-
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 - PubMed
-
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M (2006) Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38: 1289–1297 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

