Exon array analysis reveals neuroblastoma tumors have distinct alternative splicing patterns according to stage and MYCN amplification status
- PMID: 21501490
- PMCID: PMC3096898
- DOI: 10.1186/1755-8794-4-35
Exon array analysis reveals neuroblastoma tumors have distinct alternative splicing patterns according to stage and MYCN amplification status
Abstract
Background: Neuroblastoma (NB) tumors are well known for their pronounced clinical and molecular heterogeneity. The global gene expression and DNA copy number alterations have been shown to have profound differences in tumors of low or high stage and those with or without MYCN amplification. RNA splicing is an important regulatory mechanism of gene expression, and differential RNA splicing may be associated with the clinical behavior of a tumor.
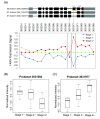
Methods: In this study, we used exon array profiling to investigate global alternative splicing pattern of 47 neuroblastoma samples in stage 1 and stage 4 with normal or amplified MYCN copy number (stage 1-, 4- and 4+). The ratio of exon-level expression to gene-level expression was used to detect alternative splicing events, while the gene-level expression was applied to characterize whole gene expression change.
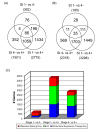
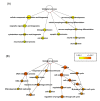
Results: Principal component analysis (PCA) demonstrated distinct splicing pattern in three groups of samples. Pairwise comparison identified genes with splicing changes and/or whole gene expression changes in high stage tumors. In stage 4- compared with stage 1- tumors, alternatively spliced candidate genes had little overlap with genes showing whole gene expression changes, and most of them were involved in different biological processes. In contrast, a larger number of genes exhibited either exon-level splicing, gene-level expression or both changes in stage 4+ versus stage 1- tumors. Those biological processes involved in stage 4- tumors were disrupted to a greater extent by both splicing and transcription regulations in stage 4+ tumors.
Conclusions: Our results demonstrated a significant role of alternative splicing in high stage neuroblastoma, and suggested a MYCN-associated splicing regulation pathway in stage 4+ tumors. The identification of differentially spliced genes and pathways in neuroblastoma tumors of different stages and molecular subtypes may be important to the understanding of cancer biology and the discovery of diagnostic markers or therapeutic targets in neuroblastoma.
Figures


References
-
- Thorsen K, Sorensen KD, Brems-Eskildsen AS, Modin C, Gaustadnes M, Hein AM, Kruhoffer M, Laurberg S, Borre M, Wang K, Brunak S, Krainer AR, Tørring N, Dyrskjøt L, Andersen CL, Orntoft TF. Alternative splicing in colon, bladder, and prostate cancer identified by exon-array analysis. Mol Cell Proteomics. 2008. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

