Analysis of anther transcriptomes to identify genes contributing to meiosis and male gametophyte development in rice
- PMID: 21554676
- PMCID: PMC3112077
- DOI: 10.1186/1471-2229-11-78
Analysis of anther transcriptomes to identify genes contributing to meiosis and male gametophyte development in rice
Abstract
Background: In flowering plants, the anther is the site of male gametophyte development. Two major events in the development of the male germline are meiosis and the asymmetric division in the male gametophyte that gives rise to the vegetative and generative cells, and the following mitotic division in the generative cell that produces two sperm cells. Anther transcriptomes have been analyzed in many plant species at progressive stages of development by using microarray and sequence-by synthesis-technologies to identify genes that regulate anther development. Here we report a comprehensive analysis of rice anther transcriptomes at four distinct stages, focusing on identifying regulatory components that contribute to male meiosis and germline development. Further, these transcriptomes have been compared with the transcriptomes of 10 stages of rice vegetative and seed development to identify genes that express specifically during anther development.
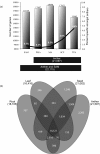
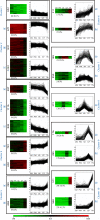
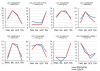
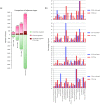
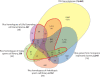
Results: Transcriptome profiling of four stages of anther development in rice including pre-meiotic (PMA), meiotic (MA), anthers at single-celled (SCP) and tri-nucleate pollen (TPA) revealed about 22,000 genes expressing in at least one of the anther developmental stages, with the highest number in MA (18,090) and the lowest (15,465) in TPA. Comparison of these transcriptome profiles to an in-house generated microarray-based transcriptomics database comprising of 10 stages/tissues of vegetative as well as reproductive development in rice resulted in the identification of 1,000 genes specifically expressed in anther stages. From this sub-set, 453 genes were specific to TPA, while 78 and 184 genes were expressed specifically in MA and SCP, respectively. The expression pattern of selected genes has been validated using real time PCR and in situ hybridizations. Gene ontology and pathway analysis of stage-specific genes revealed that those encoding transcription factors and components of protein folding, sorting and degradation pathway genes dominated in MA, whereas in TPA, those coding for cell structure and signal transduction components were in abundance. Interestingly, about 50% of the genes with anther-specific expression have not been annotated so far.
Conclusions: Not only have we provided the transcriptome constituents of four landmark stages of anther development in rice but we have also identified genes that express exclusively in these stages. It is likely that many of these candidates may therefore contribute to specific aspects of anther and/or male gametophyte development in rice. In addition, the gene sets that have been produced will assist the plant reproductive community in building a deeper understanding of underlying regulatory networks and in selecting gene candidates for functional validation.
Figures




References
-
- Yu-Jin Sun CLH, Chen Chang-Bin, Hong Ma. Regulation of Arabidopsis early anther development by putative cell-cell signalling molecules and transcriptionl regulators. Journal of Integrative Plant Biology. 2007;49:60–68. doi: 10.1111/j.1744-7909.2006.00408.x. - DOI
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

