Targeting BARD1 suppresses a Myc-dependent transcriptional program and tumor growth in pancreatic ductal adenocarcinoma
- PMID: 40096771
- PMCID: PMC11957605
- DOI: 10.1016/j.neo.2025.101152
Targeting BARD1 suppresses a Myc-dependent transcriptional program and tumor growth in pancreatic ductal adenocarcinoma
Abstract
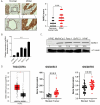
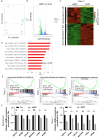
Pancreatic ductal adenocarcinoma (PDAC) remains one of the deadliest cancers demanding better and more effective therapies. BARD1 or BRCA1-Associated -Ring Domain-1 plays a pivotal role in homologous recombination repair (HRR). However, its function and the underlying molecular mechanisms in PDAC are still not fully elucidated. Here, we demonstrate that BARD1 is overexpressed in PDAC and its genetic inhibition suppresses c-Myc and disrupts c-Myc dependent transcriptional program. Mechanistically, BARD1 stabilizes c-Myc through ubiquitin-proteasome system by regulating FBXW7. Importantly, targeting BARD1 using either siRNAs or CRISPR/Cas9 deletion blocks PDAC growth in vitro and in vivo, without any signs of toxicity to mice. Using a focused drug library of 477 DNA damage response compounds, we also found that BARD1 inhibition enhances therapeutic efficacy of several clinically relevant agents (fold changes ≥4), including PARPi, in HRR proficient PDAC cells. These data uncover BARD1 as an attractive therapeutic target for HRR proficient PDAC.
Keywords: BARD1; DNA damage; PARP inhibitor; Pancreatic ductal adenocarcinoma; c-Myc.
Copyright © 2025. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

