IL-12 drives the expression of the inhibitory receptor NKG2A on human tumor-reactive CD8 T cells
- PMID: 39557863
- PMCID: PMC11574270
- DOI: 10.1038/s41467-024-54420-w
IL-12 drives the expression of the inhibitory receptor NKG2A on human tumor-reactive CD8 T cells
Abstract
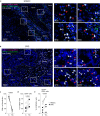
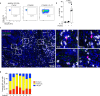
Blockade of NKG2A/HLA-E interaction is a promising strategy to unleash the anti-tumor response. Yet the role of NKG2A+ CD8 T cells in the anti-tumor response and the regulation of NKG2A expression on human tumor-infiltrating T cells are still poorly understood. Here, by performing CITE-seq on T cells derived from head and neck squamous cell carcinoma and colorectal cancer, we show that NKG2A expression is induced on CD8 T cells differentiating into cytotoxic, CD39+CD103+ double positive (DP) cells, a phenotype associated with tumor-reactive T cells. This developmental trajectory leads to TCR repertoire overlap between the NKG2A- and NKG2A+ DP CD8 T cells, suggesting shared antigen specificities. Mechanistically, IL-12 is essential for the expression of NKG2A on CD8 T cells in a CD40/CD40L- dependent manner, in conjunction with TCR stimulation. Our study thus reveals that NKG2A is induced by IL-12 on human tumor-reactive CD8 T cells exposed to a TGF-β-rich environment, highlighting an underappreciated immuno-regulatory feedback loop dependent on IL-12 stimulation.
© 2024. The Author(s).
Conflict of interest statement
Figures






References
-
- Simoni, Y. et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature557, 575–579 (2018). - PubMed
-
- Rajamanickam, V. et al. Robust antitumor immunity in a patient with metastatic colorectal cancer treated with cytotoxic regimens. Cancer Immunol. Res. 9, 602–611 (2021). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

