Genetic regulators of large-scale transcriptional signatures in cancer
- PMID: 16518402
- PMCID: PMC1435790
- DOI: 10.1038/ng1752
Genetic regulators of large-scale transcriptional signatures in cancer
Abstract
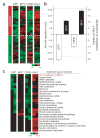
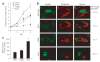
Gene expression signatures encompassing dozens to hundreds of genes have been associated with many important parameters of cancer, but mechanisms of their control are largely unknown. Here we present a method based on genetic linkage that can prospectively identify functional regulators driving large-scale transcriptional signatures in cancer. Using this method we show that the wound response signature, a poor-prognosis expression pattern of 512 genes in breast cancer, is induced by coordinate amplifications of MYC and CSN5 (also known as JAB1 or COPS5). This information enabled experimental recapitulation, functional assessment and mechanistic elucidation of the wound signature in breast epithelial cells.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests (see the
Figures





References
-
- Liu ET. Classification of cancers by expression profiling. Curr Opin Genet Dev. 2003;13:97–103. - PubMed
-
- Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- van ‘t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

