Nucleotide deficiency promotes genomic instability in early stages of cancer development
- PMID: 21529715
- PMCID: PMC3740329
- DOI: 10.1016/j.cell.2011.03.044
Nucleotide deficiency promotes genomic instability in early stages of cancer development
Abstract
Chromosomal instability in early cancer stages is caused by stress on DNA replication. The molecular basis for replication perturbation in this context is currently unknown. We studied the replication dynamics in cells in which a regulator of S phase entry and cell proliferation, the Rb-E2F pathway, is aberrantly activated. Aberrant activation of this pathway by HPV-16 E6/E7 or cyclin E oncogenes significantly decreased the cellular nucleotide levels in the newly transformed cells. Exogenously supplied nucleosides rescued the replication stress and DNA damage and dramatically decreased oncogene-induced transformation. Increased transcription of nucleotide biosynthesis genes, mediated by expressing the transcription factor c-myc, increased the nucleotide pool and also rescued the replication-induced DNA damage. Our results suggest a model for early oncogenesis in which uncoordinated activation of factors regulating cell proliferation leads to insufficient nucleotides that fail to support normal replication and genome stability.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

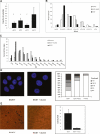

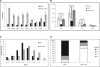

Comment in
-
Does metabolite deficiency mark oncogenic cell cycles?Cell. 2011 Apr 29;145(3):337-8. doi: 10.1016/j.cell.2011.04.003. Cell. 2011. PMID: 21529708
References
-
- Akli S, Keyomarsi K. Cyclin E and its low molecular weight forms in human cancer and as targets for cancer therapy. Cancer Biol. Ther. 2003;2(4, Suppl 1):S38–S47. - PubMed
-
- Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. - PubMed
-
- Bartkova J, Horejsí Z, Koed K, Krämer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancerbarrierinearly humantumorigenesis. Nature. 2005;434:864–870. - PubMed
-
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Onco-gene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. - PubMed
-
- Bartkova J, Hamerlik P, Stockhausen MT, Ehrmann J, Hlobilkova A, Laursen H, Kalita O, Kolar Z, Poulsen HS, Broholm H, et al. Replication stress and oxidative damage contribute to aberrant constitutive activation of DNA damage signalling in human gliomas. Oncogene. 2010;29:5095–5102. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

