The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells
- PMID: 21552207
- PMCID: PMC3116275
- DOI: 10.1038/emboj.2011.140
The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells
Abstract
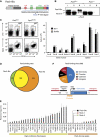
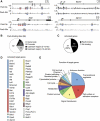
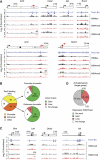
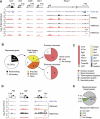
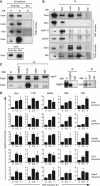
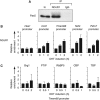
Pax5 is a critical regulator of B-cell commitment. Here, we identified direct Pax5 target genes by streptavidin-mediated ChIP-chip analysis of pro-B cells expressing in vivo biotinylated Pax5. By binding to promoters and enhancers, Pax5 directly regulates the expression of multiple transcription factor, cell surface receptor and signal transducer genes. One of the newly identified enhancers was shown by transgenic analysis to confer Pax5-dependent B-cell-specific activity to the Nedd9 gene controlling B-cell trafficking. Profiling of histone modifications in Pax5-deficient and wild-type pro-B cells demonstrated that Pax5 induces active chromatin at activated target genes, while eliminating active chromatin at repressed genes in committed pro-B cells. Pax5 rapidly induces these chromatin and transcription changes by recruiting chromatin-remodelling, histone-modifying and basal transcription factor complexes to its target genes. These data provide novel insight into the regulatory network and epigenetic regulation, by which Pax5 controls B-cell commitment.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Adams B, Dörfler P, Aguzzi A, Kozmik Z, Urbánek P, Maurer-Fogy I, Busslinger M (1992) Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev 6: 1589–1607 - PubMed
-
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ III, Gingeras TR, Schreiber SL, Lander ES (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 - PubMed
-
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

